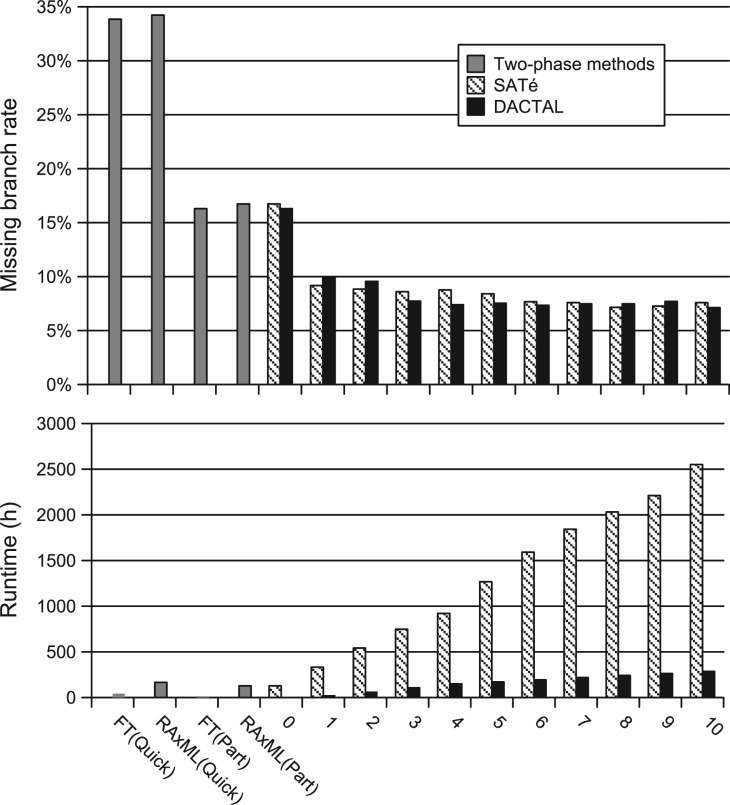
Fig. 4.
Comparisons of DACTAL and SATé iterations with two-phase methods on 16S.T dataset with 7 350 sequences. The starting trees were RAxML(Part) for SATé and FT(Part) for DACTAL. We show missing branch rates (top) and cumulative runtimes in hours (bottom); n=1 for each reported value. Iteration 0 is used to compute the starting tree for DACTAL and SATé.