Abstract
Summary: We present an updated version of the TFold software for pinpointing differentially expressed proteins in shotgun proteomics experiments. Given an FDR bound, the updated approach uses a theoretical FDR estimator to maximize the number of identifications that satisfy both a fold-change cutoff that varies with the t-test P-value as a power law and a stringency criterion that aims to detect lowly abundant proteins. The new version has yielded significant improvements in sensitivity over the previous one.
Availability: Freely available for academic use at http://pcarvalho.com/patternlab.
Contact: paulo@pcarvalho.com
1 INTRODUCTION
One of the goals of shotgun proteomics is to perform large-scale comparisons of complex protein mixtures (e.g. biological fluids or whole-cell lysates). It comprises protein digestion followed by peptide chromatographic separation online with tandem mass spectrometry (MS2) for protein identification and quantitation. There are several strategies for protein quantitation; examples are metabolic labeling (Gouw et al., 2010), chemical derivatization (Thompson et al., 2003) and label-free (Bondarenko et al., 2002).
We previously described PatternLab for Proteomics (Carvalho et al., 2010), a computational environment for analyzing shotgun proteomics data. The modules available in PatternLab serve various purposes, ranging from preparing decoy databases, to selecting and sharing reliable protein identifications (Carvalho et al., 2012), to performing gene ontology analyses (Carvalho et al., 2009), and many more. One of its most popular modules is the TFold test for pinpointing differentially expressed proteins quantitated by any of the above strategies. Briefly, in the previous version a PatternLab file containing protein quantitation data from different biological conditions would first be filtered according to specified t-test P-value and fold-change cutoffs. Then the user would interactively experiment with different fold-change cutoffs, each leading to a different list of proteins through the BH FDR estimator (Benjamini and Hochberg, 1995) for the same user-specified q-value, henceforth referred to as α. This process would ultimately lead to a fold-change cutoff that maximized the number of proteins in the list.
Even though this simple strategy has been effective in a number of occasions, imposing a fixed fold-change cutoff could discard proteins with very low P-values but not satisfying the fold-change cutoff. This seems unreasonable because, in the proteomics scenario, the magnitude of a protein's fold change need not correlate with its biological importance. In fact, molecular abundance is only one of the various factors controlling protein activity, not necessarily the most important one (Weiss et al., 2010). Here, we describe how we reformulated the TFold test to better address these limitations and increase sensitivity.
We also tackle another common problem in proteomics, namely that of dealing with lowly abundant proteins (e.g. those having low spectral counts), which usually turn out to be over-represented in complex mixtures. These proteins are more prone to getting around the barriers imposed by common statistical filters (e.g. the t-test) because they tend to artificially acquire low P-values. To exemplify, assume the following sets of spectral counts for a given protein in two biological conditions: {1, 1, 1} and {5, 6, 5}. Notably, in this extreme example, there is a considerable fold change while the P-value is artificially low due to the lack of variation. In our experience, it is much more likely that this protein is a false-positive than another having significantly higher spectral counts but with a higher P-value as well. Our updated TFold approach introduces a method to quickly highlight (and separate) proteins such as the one here exemplified. For these proteins, additional experimentation is recommended before ascertaining their status as differentially expressed.
2 VARIABLE FOLD-CHANGE CUTOFF
The centerpiece of our fold-change reformulation is to allow the fold-change cutoff for a protein to be given as a function of its t-test P-value. In order to address the issues raised above, a larger than 1 fold-change cutoff must decrease as the P-value decreases; likewise, a smaller than 1 fold-change cutoff must increase as the P-value decreases. We postulate a power-law functional form for some exponent z>0, thence the fold−change cutoff is proportional to pz in the former case and to p−z in the latter, where p is the P-value in question. We resolve the proportionality constant by imposing a fold-change cutoff of 1 when p is the lowest P-value for the proteins in the data set, here denoted by pmin. A protein of P-value p is then rejected (i.e. declared not differentially expressed) if its fold change lies between (pmin/p)z and (p/pmin)z.
Thus, in the revised version of the TFold test, one core operation is to filter the proteins according to the P-value-dependent fold-change cutoffs that result from a fixed value of the exponent z. Building on this operation, our software automatically maximizes the list of identified proteins by varying the value of z given α (decreasing the value of z tightens the fold-change interval for protein rejection). Thus, z works as a fold-change stringency parameter (F-stringency).
3 HANDLING LOWLY ABUNDANT PROTEINS
Another core operation is to filter and highlight proteins that would be considered differentially expressed according to α and the fold-change cutoff function but still have a high chance of being false positives. This occurs mainly in the case of lowly abundant proteins, as their quantitation values are often compromised. Briefly, we begin by assuming that the quantitation value of a randomly chosen quantitated protein, regardless of biological condition, is exponentially distributed with parameter λ which implies that both mean and standard deviation are given by 1/λ. As is well-known, the maximum-likelihood estimates of 1/λ is the sample mean of all proteins' quantitation values (to which a protein contributes as many times as there are biological conditions in which it appears).
This given, a new parameter, called λ -stringency (L-stringency), is introduced to pinpoint each problematic protein. This is done by specifying a fraction of 1/λ below which the sample mean of that protein in a certain biological condition is too low for it to qualify as differentially expressed in that condition. We exemplify the use of L-stringency as follows. Suppose that the sets {2, 2, 2} and {7, 8, 7} of spectral counts were acquired for a certain protein in two biological conditions and that 1/λ=2. Choosing an L-stringency of 0.4 disqualifies the protein as differentially expressed in the second biological condition relative to the first, since (7+8+7)/3 <0.4×20. We recommend this L-stringency of 0.4, but it should be evaluated by the user on a case-by-case basis.
4 RESULTS
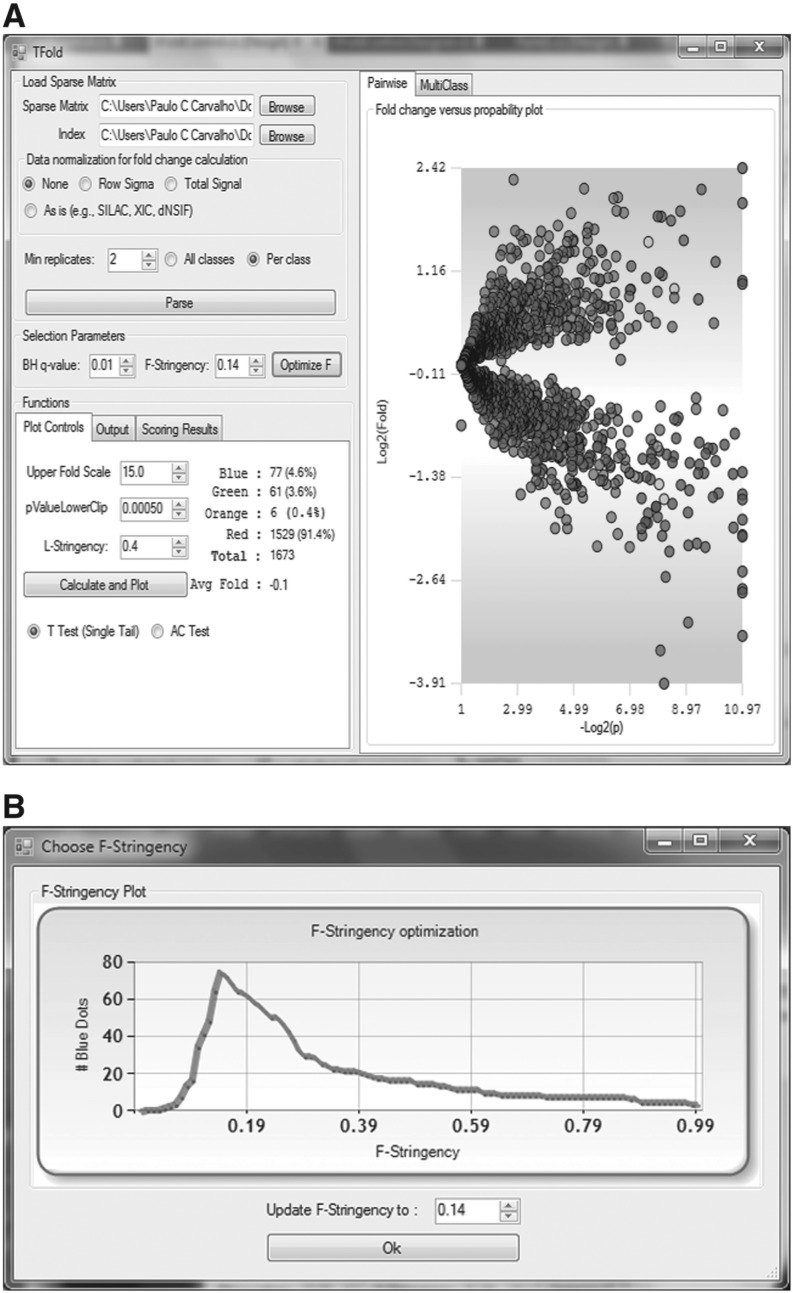
The effectiveness of our updated algorithm is exemplified on the data set provided by Fischer et al. (2011) comparing A172 cancer cells to those (A172R) resistant to Perillyl Alcohol (POH), a naturally occurring chemotherapeutic agent that induces apoptosis. The authors provide a list of protein identifications quantitated by spectral counting and satisfying a 1% FDR for their identifications according to DTASelect (Cociorva et al., 2006). In the original analysis, the authors converged to a fixed fold-change cutoff of 2.5, resulting in 57 differentially expressed proteins for a P-value cutoff of 0.01 and α=0.01. The new algorithm converged to z=0.14, pinpointing 77 differentially expressed proteins for the same α, plus six proteins satisfying the same criteria but reported in a separate list as having been marked by the L-stringency filter, thus suggesting the need for further experimentation. Figure 1 is a snapshot of Pattern Lab's graphical user interface during analysis.
Fig. 1.
Panel A shows the TFold graphical user interface and the results of our updated analysis relative to Fischer et al. (2011). Each protein is mapped as a dot on the plot according to its –log2(P-value) (x-axis) and log2(fold change) (y-axis). Red dots are proteins that satisfy neither the fold-change cutoff nor the FDR cutoff α. Green dots are those that satisfy the fold-change cutoff but not α. Orange dots are those that satisfy both the fold-change cutoff and α but are lowly abundant proteins and thus require further experimentation to certify their differential expression. Blue dots, finally, are proteins that satisfy all statistical filters. Note that the bounds separating green dots from red do not correspond to a single fold-change cutoff (a ‘horizontal’ line) as in the previous approach. Instead, separation is achieved through several cutoffs along a ‘conical’ region (due to the logarithms), one cutoff for each P-value, with vertex at P-value =pmin and Fold change = 1. However, as a consequence of the BH FDR estimator, we have a single P-value cutoff (a vertical line) separating blue and orange dots from green and red in both approaches. Panel B is automatically displayed once the Optimize button is pressed. Given α, it plots the distribution of blue dots as a function of the F-stringency parameter z
An example of a protein that was only marked as differentially expressed by the revised method is glyceraldehyde-3-phosphate dehydrogenase (GAPDH; IPI00219018.7). Identifying this protein is important because we hypothesize that it could be related to the initiation of apoptosis when the cell's medium contains POH. The sets of spectral counts for this protein were {117, 71, 141} for the wild-type and {271, 255, 240} for the resistant cell culture, but further discussions on any of the corresponding biological interpretations are beyond the scope of this manuscript.
5 FINAL CONSIDERATIONS
Computational approaches for pinpointing differentially expressed proteins can, at best, provide a list of putative biomarkers that could ultimately aid in the understanding of pathology or be specific to a biological condition. Further validation is to be carried out in different cohorts.
The application of the F-stringency parameter prior to using the BH FDR estimator eliminates hypotheses (i.e. putative differentially expressed proteins) that are most likely false positives. Applying the L-stringency parameter also prior to using the BH FDR estimator has the potential of further eliminating the likely false positives related to some of the lowly abundant proteins. The final, reduced list to be evaluated by the BH FDR estimator maximizes the TFold test sensitivity, as the BH FDR estimator penalizes the number of hypotheses tested by including it in the denominator of its equation.
To summarize, we have described an algorithm for maximizing the number of differentially expressed protein candidates. It optimizes a fold-change cutoff that varies with the proteins' P-values as a power law and provides a means for highlighting (and separating) lowly abundant proteins prior to filtering by the BH FDR estimator. This algorithm resulted in a significant increase in sensitivity when compared to its previous version.
ACKNOWLEDGEMENT
The authors thank Prof. Gilberto Barbosa Domont for critically reviewing the manuscript.
Funding: PCC and VCB were funded by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) /Fiocruz 30/2006, Programa de Desenolvimento Tecnológico de Insumos para a Saúde (PDTIS), Conselho Nacional de Pesquisa (CNPq,) Fundaç ao de Amparo a Pesquisa do Rio de Janeiro (FAPERJ BBP). JRY was funded by the National Institutes of Health (NIH) [P41 RR011823, P41 GM130533, 5R01 MH067880].
Conflict of Interest: none declared.
REFERENCES
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- Bondarenko P.V., et al. Identification and relative quantitation of protein mixtures by enzymatic digestion followed by capillary reversed-phase liquid chromatography-tandem mass spectrometry. Anal. Chem. 2002;74:4741–4749. doi: 10.1021/ac0256991. [DOI] [PubMed] [Google Scholar]
- Carvalho P.C., et al. GO Explorer: a gene-ontology tool to aid in the interpretation of shotgun proteomics data. Proteome Sci. 2009;7:6. doi: 10.1186/1477-5956-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P.C., et al. Analyzing shotgun proteomic data with PatternLab for proteomics. Curr. Protoc. Bioinformatics. 2010 doi: 10.1002/0471250953.bi1313s30. Chapter 13, Unit 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P.C., et al. Search engine processor: filtering and organizing PSMs. Proteomics. 2012;12:944–949. doi: 10.1002/pmic.201100529. doi: 10.1002/pmic.201100529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cociorva D., et al. Validation of tandem mass spectrometry database search results using DTASelect. Curr. Protoc. Bioinformatics. 2006:13.4.1–13.4.14. doi: 10.1002/0471250953.bi1304s16. Chapter 13. [DOI] [PubMed] [Google Scholar]
- Fischer J.S.G., et al. Chemo-resistant protein expression pattern of glioblastoma cells (A172) to perillyl alcohol. J. Proteome Res. 2011;10:153–160. doi: 10.1021/pr100677g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouw J.W., et al. Quantitative proteomics by metabolic labeling of model organisms. Mol. Cell Proteom. 2010;9:11–24. doi: 10.1074/mcp.R900001-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A., et al. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 2003;75:1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- Weiss M., et al. Shotgun proteomics data from multiple organisms reveals remarkable quantitative conservation of the eukaryotic core proteome. Proteomics. 2010;10:1297–1306. doi: 10.1002/pmic.200900414. [DOI] [PubMed] [Google Scholar]