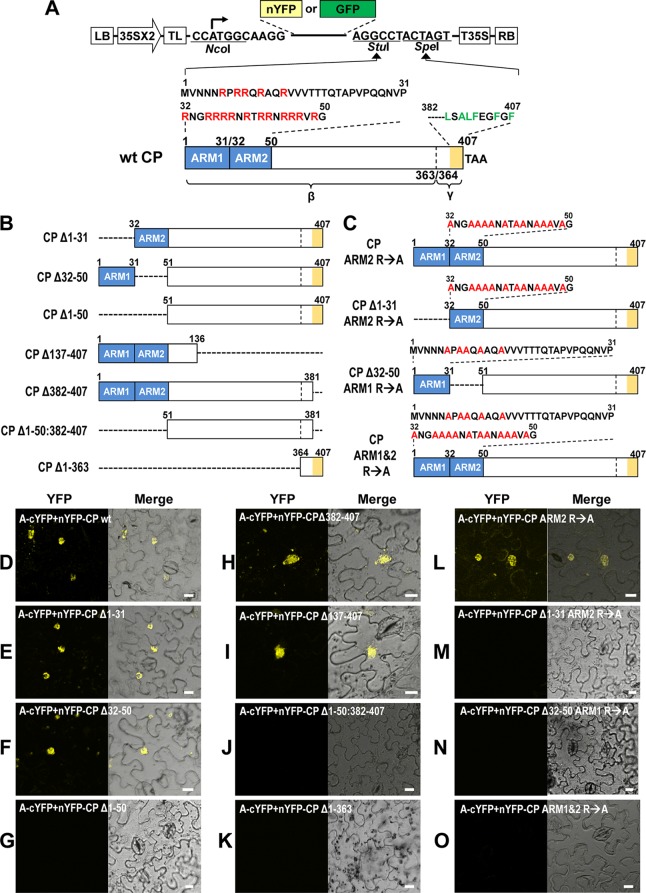
Fig 5.
Identification of CP domains required for the interaction with protein A. (A) Schematic representation of CP variant containing deletions and amino acid substitutions. Characteristic features of a binary vector, as described in the Fig. 1 legend, containing either nYFP or GFP is shown. A wt FHV CP (wt CP) of 407 amino acids is shown. The N-terminal region from amino acids 1 to 50 is divided into two regions designated ARM1 (amino acids 1 to 30 containing 5 arginine residues indicated in red font) and ARM2 (amino acids 32 to 50 containing 12 arginine residues indicated in red font) and are indicated by blue boxes. The region spanning mature β-subunit (amino acids 1 to 363) and the γ-subunit (amino acids 364 to 407) of the CP is indicated. The C terminus of the CP located between amino acids 382 and 407, indicated by orange shading, contains a high proportion of hydrophobic amino acid residues. (B) Dashed lines represent the extent of engineered deletions in the CP ORF. (C) Arginine residues located in ARM1, ARM2, or both were substituted with alanine residues (indicated in red font). nYFP fusions for each CP mutant (shown in Fig. 5A to C) were constructed by using the binary vector shown in Fig. 1. (D to O) Each nYFP-CP mutant was coexpressed with A-cYFP in N. benthamiana leaves by agroinfiltration. The reconstructed YFP signals were observed in the epidermal cells using confocal microscopy at 3 dpi. Images emitting yellow fluorescence by reconstituted YFP are shown to the left, and images merged with those taken under the transmitted-light mode are shown to the right. Bar, 15 μm.