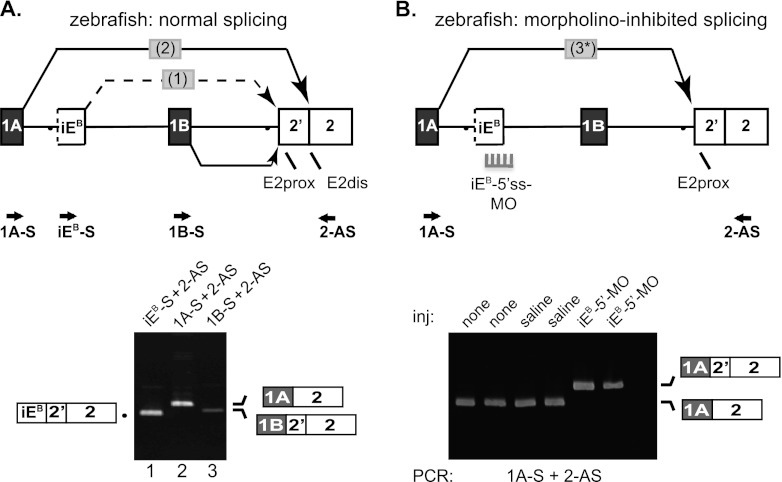
Fig 6.
Analysis of intrasplicing in the zebrafish 4.1B gene. (A) (Top) 5′ structure of the zebrafish 4.1B gene, including the putative intraexon and two nested splicing reactions predicted to be essential for E1A to E2dis splicing. Locations of PCR primers are indicated. Bottom: RT-PCR analysis of normal zebrafish 4.1B transcripts. Lane 1, intermediate RNA product obtained using primers iEB-S and 2-AS; lane 2, E1A-E2dis splicing revealed using primers 1A-S and 2-AS; lane 3, E1B-E2prox splicing demonstrated using primers 1BF and 2R. B. Top: Model for switch in splice acceptor site usage induced in vivo by inhibition of intraexon function. Splice-blocking MO directed against the intraexon 5′ splice site (5′ss-MO) inhibited splicing events 1 and 2, leading to direct splicing of E1A to E2prox in a single aberrant splicing event 3. Bottom: RT-PCR analysis of duplicate experiments using primers 1A-S and 2-AS to amplify zebrafish RNA prepared from uninjected embryos (none); embryos injected with 0.2 M KCl buffer alone (saline); or injected with 2 or 10 ng of 5′ss-MO against the 4.1B intraexon (iEB-5′-MO).