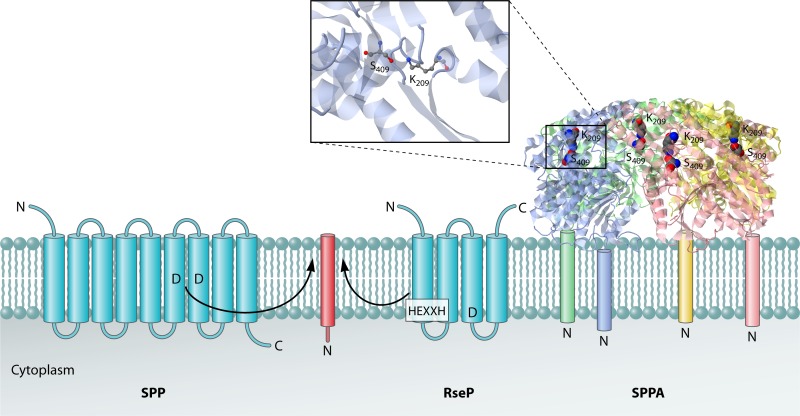
Fig 4.
Cleavage of signal peptides by signal peptide hydrolases. The eukaryotic signal peptide hydrolase/peptidase SPP is an aspartic acid protease with catalytic Asp residues located within the plane of the membrane. The RseP protease, which seems to function as a general bacterial signal peptide hydrolase, is a metalloprotease with an intramembrane catalytic site facing the cytoplasmic side of the membrane. The HEXXH motif that binds the catalytic Zn2+ ion is indicated. The bacterial signal peptide peptidase SPPA is a homotetramer with catalytic Ser-Lys dyads in domains that are juxtapositioned to the extracytoplasmic membrane surface. Transmembrane helices of SPP and RseP are depicted as blue barrels, and the substrate helix is depicted as a red barrel. Furthermore, the four subunits of the SPPA complex are depicted in blue, green, red, and yellow. A zoomed-in view of the active site residues of SPPA is shown. The PDB structure 3BF0 and the program JMol were used to generate the 3D structure image of SPPA. The proposed position of the SPPA N-terminal transmembrane segment for each monomer (which was missing from the construct used to the solve the structure) is shown schematically. The locations of the N and C termini of the signal peptide hydrolases and their substrates are indicated.