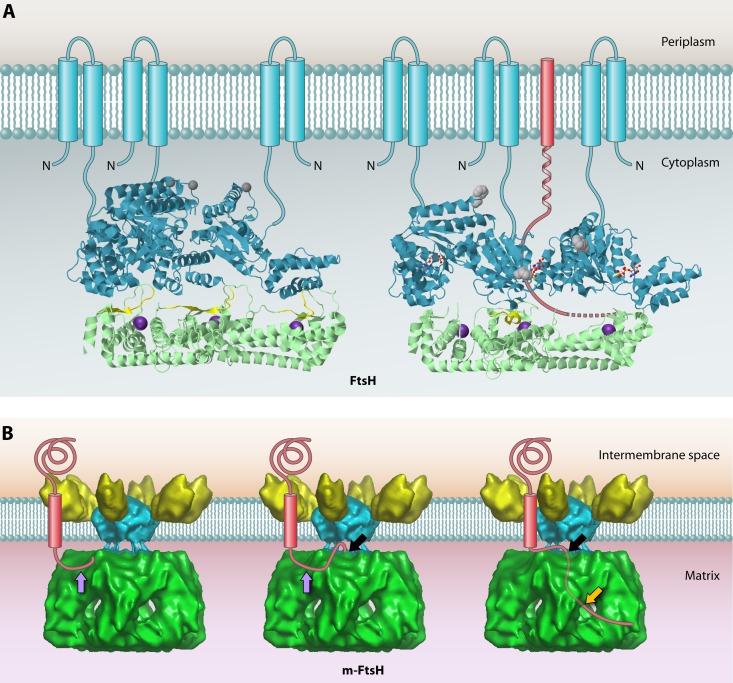
Fig 6.
Structures of bacterial FtsH and the homologous m-AAA protease in the inner mitochondrial membrane. (A) Structures of the cytoplasmic domains of apo-FtsH (left) and ADP-FtsH (right). The AAA domains are shown in cyan, and protease domains are shown in green. To give a clear inside view of the hexamer chamber, only 3 subunits, forming half of the holoenzyme, are shown. Amino acids 450 to 460 are proposed to form an active site switch (highlighted in yellow). These residues change from a β-sheet conformation in the apoprotein to an α-helical conformation in the ADP-bound form, which closes the proteolytic site of the corresponding subunit. The inward movement of this AAA domain opens the substrate tunnel, and the substrate polypeptide chain (red) is pulled through the chamber toward the open proteolytic site of the adjacent subunit. Phe234 at the substrate binding pore is shown in gray. The proposed positions of the N-terminal transmembrane segments of each monomer (which were missing from the construct used to the solve the structure) are shown schematically. Bound ADP is shown as a ball-and-stick model. The Zn2+ ion at the proteolytic site is shown as an enlarged purple sphere. The apo-FtsH structure was obtained from PDB accession number 3KDS, and that of ADP-FtsH was obtained from PDB accession number 2CEA. JMol was used to generate the 3D structure images. (B) Proposed mode of action of the mitochondrial m-AAA protease based on the cryo-electron microscopy (cryo-EM) structure. The structure data were obtained from EMDataBank (accession number 1712), and the Astex Viewer was used to generate the image. From left to right, the model depicts subsequent stages in substrate degradation. First, an unfolded terminal peptide of the substrate binds the surface of the AAA domain at the initial contact site (purple arrow). Subsequently, the unfolded peptide is transferred to the secondary binding site (black arrow), where it enters the AAA ring through the center pore. Lastly, the substrate is degraded in the chamber of the protease domain, and the resulting degradation products are released from the side pores (orange arrow).