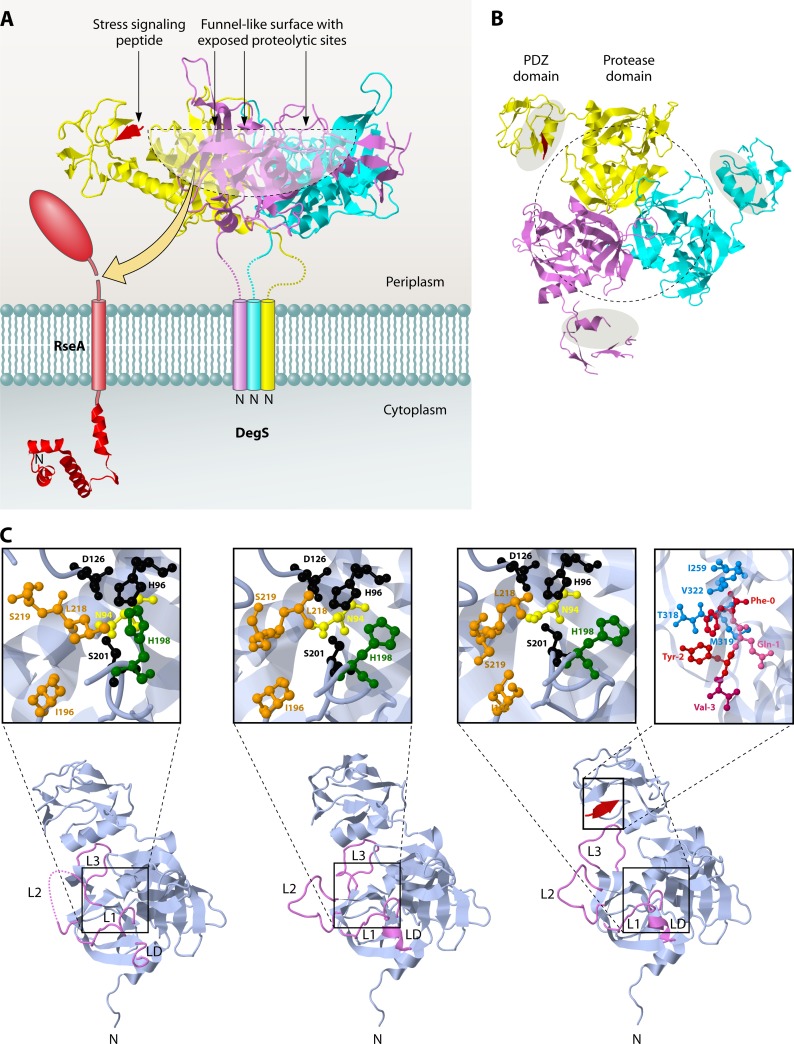
Fig 8.
Trimeric structure of the sheddase DegS. (A) Side view of DegS. (B) View from the periplasmic side. The protease domains in the center surround a funnel-like exposed surface where the proteolytic sites are located (outlined by a black dotted line). The PDZ domains that contain the peptide-binding grooves (gray shaded areas in panel B) are located on the outside of the DegS trimer. They are connected to the protease domains at their C termini. A peptide that is bound to the PDZ domain in one of the subunits is colored red. The structure data were obtained from PDB accession number 1SOZ, and JMol was used to generate the images. (C) Monomer structures of DegS in the peptide-free inactive (left), intermediate (middle), and peptide-bound active (right) states. The L1, L2, L3, and LD loops are highlighted in violet. In the intermediate form, without a substrate, L3 retreats from the substrate docking position (active form) to the inactive conformation, while L1, L2, and LD, located at the active site center, still adopt the active conformation. The active sites are shown in enlarged insets for all structures. The catalytic residues Ser201, His96, and Asp126 are shown in black. Residues forming the S1 pocket are shown in orange. These residues are reorganized upon substrate binding (compare the inactive and active forms). N94 (yellow) and H198 (green), which interfere with the catalytic triad and destabilize the oxyanion hole, as shown in the inactive form, move away in the active form. The stress signaling peptide binding site in the PDZ domain for DegS in the proteolytic active state is also shown in an enlarged inset. The peptide is colored red, and residues forming the hydrophobic pocket surrounding the Phe0 position are colored blue. Structure data were obtained from PDB, using accession numbers 1SOT (inactive state), 1VCW (intermediate state), and 1SOZ (active state). JMol was used to construct the images.