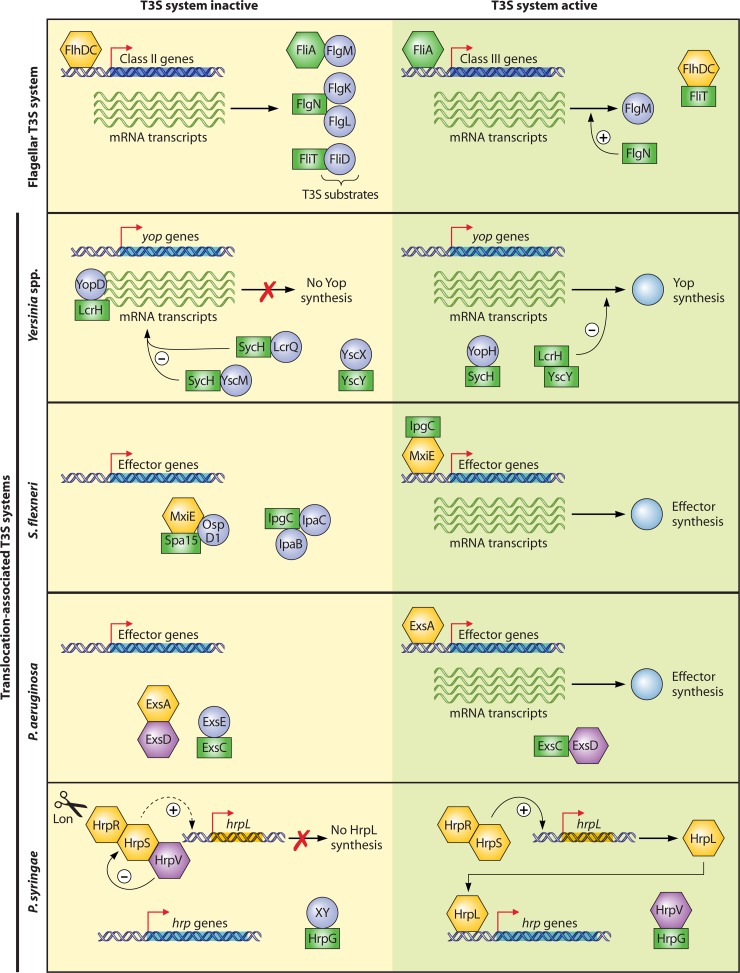
Fig 7.
Schematic representation of the regulatory mechanisms that control T3S gene expression in flagellar and translocation-associated T3S systems. The expression of effector genes or genes encoding substrates and components of the flagellar T3S system is controlled by transcriptional regulators and regulatory chaperones and depends on the activity of the T3S system. Flagellar gene expression is regulated by the transcriptional activators FlhDC, which are encoded by class I genes and activate the expression of class II genes. Class II gene products include the anti-σ28 factor FlgM, the hook-filament junction proteins FlgK and FlgL, and the filament cap protein FliD, which bind to the corresponding chaperones FliA, FlgN, and FliT, respectively. The secretion of FlgM leads to the release of the σ28 factor FliA, which activates the expression of class III genes. The liberated T3S chaperone FliT binds to FlhC and thus inhibits the expression of class II genes, whereas FlgN positively regulates the translation of flgM class III mRNA. In Yersinia spp., translation of yop mRNAs is suppressed by a YopD-LcrH complex when the T3S system is inactive. LcrQ-SycH and YscM-SycH complexes might act as additional repressors. Activation of Yop secretion leads to relief of the YopD-LcrH-mediated repression of yop mRNA translation and the liberation of SycH and LcrH upon secretion of YopD. The SycH chaperone presumably promotes YopH secretion after its release from the secreted regulator YscM. LcrH might suppress yop gene expression when bound to the T3S chaperone YscY. Effector gene expression in S. flexneri depends on the transcriptional activator MxiE and its coactivator, IpgC. Upon activation of T3S, MxiE and IpgC are released from their secreted interaction partners, i.e., Spa15, OpsD1, IpaC, and IpaD, and can activate effector gene expression. In P. aeruginosa, effector gene expression is controlled by the activator ExsA, which interacts with the antiactivator ExsD when the T3S system is inactive. The T3S chaperone ExsC, which binds to the T3S substrate ExsE, acts as an antiantiactivator when released from ExsE after activation of the T3S system. The interaction of ExsC with ExsD leads to the liberation of ExsA, which subsequently activates effector gene expression. In the plant-pathogenic bacterium P. syringae, expression of hrp (hypersensitive response and pathogenicity) genes that encode the T3S system is induced by HrpR, HrpS, and HrpL. HrpR and HrpS interact with each other and activate the expression of the alternative sigma factor HrpL, which binds to the promoters of hrp genes. HrpR-, HrpS-, and HrpL-dependent activation of hrp gene expression is counteracted by the negative regulator HrpV, which interacts with HrpS, and by the Lon protease, which degrades HrpR. Under T3S-inducing conditions, HrpV interacts with the chaperone-like protein HrpG, which interferes with the negative regulatory activity of HrpV and might also bind to a secreted but so far unknown interaction partner (XY). T3S chaperones are represented in green, and T3S substrates are represented by circles. Transcriptional regulators are depicted in yellow.