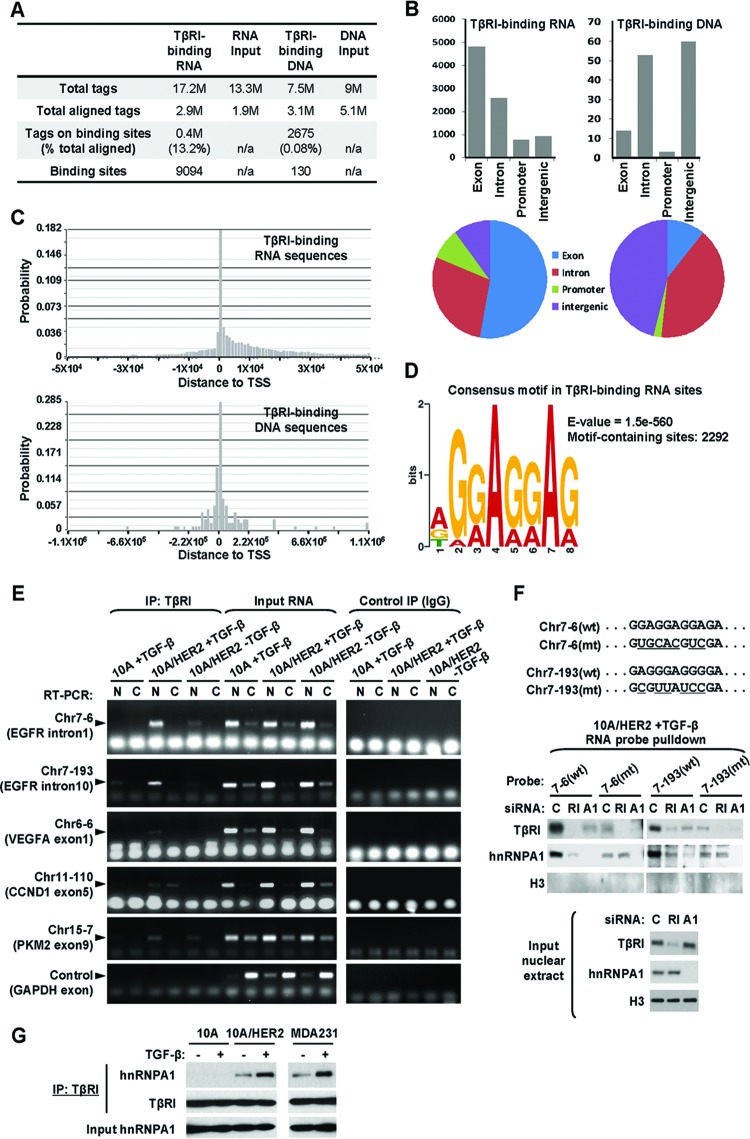
Fig 5.
Identification of TβRI-binding RNA sequences. (A) Summary of the total tags, aligned tags, tags on binding sites, and binding sites detected from ChIP or RIP deep sequencing. (B) Regional distributions of identified TβRI-binding RNA and DNA sequences. (C) Summarized locations of identified TβRI-binding RNA and DNA sequences relative to transcription start sites (TSS). (D) Consensus motif identified from TβRI-binding RNA sequences with the most significant E value, the probability of finding an equally well-conserved pattern in random sequences. (E) Confirmation of selected TβRI-binding RNA sequences using RIP followed by RT-PCR. The nuclear and cytoplasmic fractions of cells that were treated as indicated were subjected to RIP using HA tag antibody or normal IgG (as a negative control). Input RNA was used as a positive control for RT-PCR. Primer pairs specific for each selected TβRI-binding site or GAPDH (as a negative control) were used for fragment detection. (F) Pulldown assays were carried out using biotinylated RNA probes with wild-type (wt) or mutated (mt) sequences as indicated. MCF10A/HER2 cells were transfected with siRNAs of TβRI (RI), hnRNP A1 (A1), or control sequence (C) and treated with TGF-β for 0.5 h before nuclear extracts were prepared and incubated with each RNA probe. After pulldown with streptavidin beads, the eluates as well as the input nuclear extracts were analyzed by Western blotting using the indicated antibodies. (G) Co-IP assay was carried out using TβRI antibody in cells treated with TGF-β for 0.5 h or left untreated. The precipitates as well as the input lysates were analyzed by Western blotting as indicated.