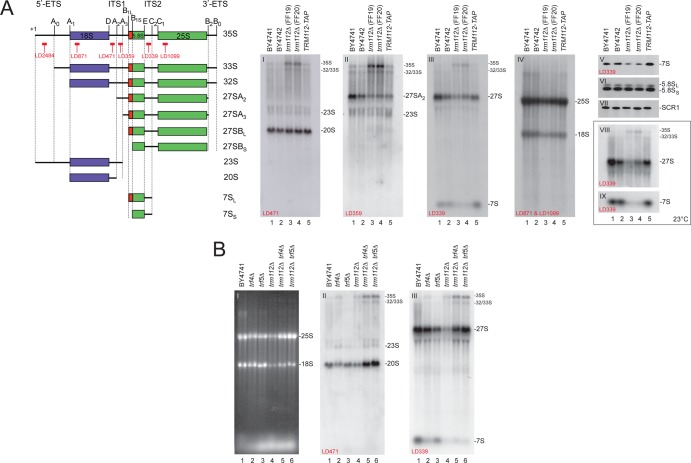
Fig 2.
Trm112 is required for efficient pre-rRNA processing, and the absence of Trm112 activates a 3′→5′ nucleolar surveillance pathway which targets preribosomes for degradation. (A) In trm112Δ cells, early pre-rRNA processing is inhibited and precursors to large subunit rRNA are unstable. (Left) Schematics depicting the pre-rRNA intermediates and probes used. (Right) Total RNA extracted from the indicated strains grown to mid-log phase in YPDA at 30°C (gels I to VII) or 23°C (gels VIII and IX) was separated on agarose-formaldehyde gels, transferred to a nylon membrane, and hybridized with a set of oligonucleotide probes in Northern blots, as indicated on the gels. Gel VI was hybridized with LD915 and gel VII with LD1290. (B) In trm112Δ cells, pre-rRNA precursors are turned over by TRAMP-dependent nucleolar surveillance. Information is the same as for panel A. Cells were grown at 30°C. Gel I, ethidium bromide staining.