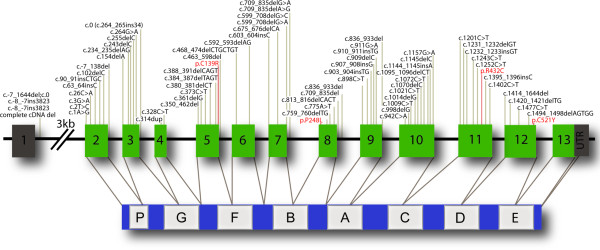
Figure 1.
Overview of pathogenic loss-of-function mutations and potential pathogenic missense progranulin mutations in frontotemporal lobar degeneration. Mutations in the progranulin gene (GRN) are a common cause of frontotemporal lobar degeneration with transactivation response DNA binding protein molecular weight of 43 kDa pathology (FTLD-TDP). Schematic representation of the genomic structure of GRN (top) and progranulin protein (PGRN) (bottom). The gray numbered boxes in the genomic structure indicate the noncoding exon 1 and the 3' untranslated region; the green numbered boxes indicate coding exons 2 to 13. The white lettered boxes in PGRN refer to the individual granulin domains. A total of 70 different loss-of-function mutations scattered over all GRN exons, except exon 13, have been reported. These mutations are indicated with vertical brown lines on the GRN exons and listed above the exons with their cDNA numbering relative to the largest GRN transcript (GenBank accession number NM_002087.2). Four potential pathogenic missense mutations are indicated in red with their protein numbering according to the largest PGRN isoform GenPept accession number NP_002078.1).