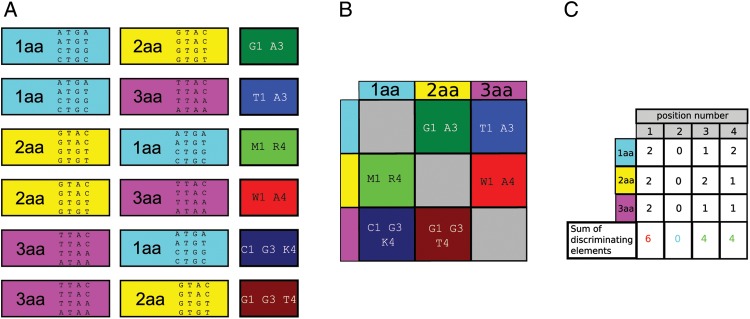
Figure 1.
Illustration of calculating the AEV using short artificial sequences. (A) Identifying DEs using an artificial set of short sequences belonging to three amino acid identity sets. Identities are labelled as 1aa, 2aa and 3aa and are highlighted as cyan, yellow and magenta, respectively. Each identity set contains four short, tetramer sequences. The calculated DE is either one base or a combination of bases and it is labelled by IUPAC codes of bases or base sets. Each step of the DE-generating algorithm is explained in Section 2. (B) The calculated DE for each identity pairs. Note that the DE-generating relationship is non-symmetrical, i.e. the 1aa vs. 2aa pair has a DE different from that of the 2aa vs. 1aa pair. (C) Positional summation of DE. Positional sum of the DE values (shown in the lowest row) provides the number of pairwise discriminations provided by the given position. The sums of the DE values are input data for calculating the AEVs). AEV is generated by dividing the positional sum of DE with the number of the identities (which in a real case is 20 while in this didactic case 3). Formalism and more detailed description are provided in Section 2. This figure can be viewed in colour online.