Abstract
The mitochondria and chloroplasts in plant cells are originated from bacterial endosymbioses, and they still replicate their own genome and divide in a similar manner as their ancestors did. It is thus likely that the organelle transcription is coordinated with its proliferation cycle. However, this possibility has not extensively been explored to date, because in most plant cells there are many mitochondria and chloroplasts that proliferate asynchronously. It is generally believed that the gene transfer from the organellar to nuclear genome has enabled nuclear control of the organelle functions during the evolution of eukaryotic plant cells. Nevertheless, no significant relationship has been reported between the organelle transcriptome and the host cell cycle even in Chlamydomonas reinhardtii. While the organelle proliferation cycle is not coordinated with the cell cycle in vascular plants, in the unicellular red alga Cyanidioschyzon merolae that contains only one mitochondrion, one chloroplast, and one nucleus per cell, each of the organelles is known to proliferate at a specific phase of the cell cycle. Here, we show that the expression of most of the organelle genes is highly coordinated with the cell cycle phases as well as with light regimes in clustering analyses. In addition, a strong correlation was observed between the gene expression profiles in the mitochondrion and chloroplast, resulting in the identification of a network of functionally related genes that are co-expressed during organelle proliferation.
Keywords: cell cycle, clustering analysis, Cyanidioschyzon, organelle, transcriptome
1. Introduction
The presence of organelle genomes in mitochondria and chloroplasts in plant cells is explained by the widely accepted endosymbiotic theory. According to this theory, mitochondria and chloroplasts originated from endosymbioses of α-proteobacteria-like and cyanobacteria-like bacteria, respectively.1,2 At the initial stage of such symbioses, the host cell would have evolved some mechanism to coordinate metabolism and proliferation of the endosymbionts to control their growth. Subsequently, a number of genes encoding proteins for organelle functions have been transferred to the nuclear genome, and the host cells have gained the ability to dominate most organelle processes by controlling the proteins involved. On the other hand, mitochondria and chloroplasts have retained the ability to undergo certain proliferation processes such as DNA replication and division. These processes require coordination of events, and thus, these organelles must control their own transcriptomes to some degree. However, the cycle-dependence of gene expression during proliferation and the relevant regulatory mechanisms are not well understood for organelles, because in most eukaryotic cells there are numerous organelles with asynchronous cycles of proliferation.
Cyanidioschyzon merolae is a thermo-acidophilic unicellular red alga isolated from an Italian volcanic hot spring.3 This organism has an extremely simple cell structure, with one nucleus, one mitochondrion, and one chloroplast. These three genome-containing organelles replicate only once at a specific phase of the cell cycle.4,5 Thus, this organism is an ideal experimental system to clarify the dependency of gene expression in the mitochondrion and the chloroplast on the proliferation cycle. Recently, the full nucleotide sequences of these three genomes were determined.6–9 Phylogenetic analyses of photosynthetic genes suggest that C. merolae is one of the most primitive photosynthetic eukaryotes, and that it diverged just after the monophyletic origin of plastids.10 The genome project also clarified that this alga has a minimally redundant small genome.8,9 Thus, it is a model eukaryote for studies on the basic architecture of eukaryotic cells based on interactions of organelles.4,11–14 Here, we report on the relationship between the replication cycle and the organelle transcriptomes in C. merolae.
Most studies on transcription of the chloroplast genome have been performed in vascular plants. Their chloroplast DNA is ∼150 kbp in length, and contains >120 genes encoding rRNAs, tRNAs, and proteins.15,16 These genes are transcribed by the cyanobacteria-type plastid-encoded plastid RNA polymerase (PEP). Sigma, which is the essential subunit for initiation of PEP transcription, is encoded by the nuclear genome, and often exists in multiple forms (e.g. six in Arabidopsis). These multiple forms are responsible for the PEP selectivity of promoter activation. Also involved in this activation is a nuclear-encoded plastid RNA polymerase (NEP), which is similar to a T7-type bacteriophage RNA polymerase. This NEP apparently evolved from the nuclear-encoded mitochondrial RNA polymerase.17,18 The nuclear genome of vascular plants encodes multiple genes for NEP-type RNA polymerases, and they are targeted to mitochondria, chloroplasts, or both. Sequential actions of these two RNA polymerases are thought to be essential for chloroplast differentiation.19,20 Genes are usually transcribed as operons, >60 of which have been described in the tobacco chloroplast genome.21 These operons tend to be processed and modified at a later point after transcription, when they are processed into mature transcripts competent for translation.22–24 The number of genes encoded in mitochondrial genomes differs among organisms. However, the transcriptional machinery is always composed of NEP-type enzymes associated with other accessory proteins.25 This system appears to be rather simple compared with those of chloroplasts.
Compared with higher plants, there is far less information available for organelle transcriptional machineries in red algae. However, it is likely that the organelle transcription machineries in C. merolae are far simpler than those in vascular plants. As revealed by bioinformatic analyses (http://www.cbs.dtu.dk/services/TargetP/), a unique NEP-type gene encoded by the nuclear genome of C. merolae is predicted to localize in the mitochondrion. This RNA polymerase is likely responsible for the transcription of 62 mitochondrial genes. PEP core subunits, α, β, and β′, are encoded by the chloroplast genome, and in addition, four sigma subunits for PEP are encoded by the nuclear genome. Besides the RNA polymerase subunits, the chloroplast genome also encodes four bacteria-type transcription factors, which were likely inherited from the ancestral cyanobacterium. These transcription factors likely sustain the organelle's autonomous transcriptional regulation in response to environmental changes.7,26
In this study, we examined the relationship between the organelle transcriptome and the cell cycle using a custom-made microarray covering two organelle genomes of C. merolae. Previously, we constructed a microarray covering the chloroplast genome, and analysed light-responsive changes of the transcriptome.26 Here, we show that there is coordinated gene expression between the chloroplast and the mitochondrion, and that the expressions of genes of both organelles are regulated both by light and by the cell cycle. In addition, a correlation clustering analysis revealed that functionally related genes show very similar expression patterns.
2. Materials and methods
2.1. Algal material and culture conditions
Cells of C. merolae 10D were cultured and their growth synchronized as described previously,27 with minor modifications. For synchronization, cells were first cultured in the MA2 medium,28 in which the concentrations of all constituents with the exception of CaCl2, FeCl3, and EDTA (disodium salt) were doubled relative to the original MA medium.29 When the optical density at 750 nm had reached 10, cells were diluted to yield an optical density of 0.5 and were cultivated under a 18 h-dark/6 h-light (150 μmol photons m−2 s−1) cycle at 42°C. Cultures were bubbled with 2% CO2 in air. Experiments with synchronized cultures were initiated at the end of the first dark period (1D18 in Supplementary Fig. S2).
2.2. RNA extraction and northern analysis
Cyanidioschyzon merolae cells were harvested by centrifugation (3000 g, 4°C, 5 min) and stored at −80°C until use. Total RNA was extracted as described previously.30 Aliquots of total RNA (5 μg) were fractioned on a 1% agarose gel containing 0.4 M formaldehyde. Probes were prepared by PCR amplification using specific primers designed from within each open reading frame. Each PCR fragment was cloned into the pT7blue vector (Novagen, Inc., Madison, WI), and then used as a probe after digestion at a suitable restriction site located in the multi-cloning site of the plasmid. Probes were labeled with alkali phosphatase using the AlkPhos direct labeling kit (GE healthcare, Giles, UK) and were hybridized to RNAs blotted onto a positively charged nylon membrane. After incubation for 16 h at 55°C and suitable washing of the membrane, hybridization signals were detected using an Image Analyzer LAS-3000 (Fuji film, Tokyo, Japan).
2.3. DNA microarray and correlation clustering analysis
We designed and constructed oligonucleotide-based microarrays that included all organelle genome-encoded open reading frames (ORFs). To synthesize dye-labeled probes for microarray analysis, we used the amino allyl RNA (aRNA) synthesis kit (ver. 2 high yield type; Sigma). Antisense cDNA was synthesized from 5 μg of total RNA by a reverse transcription reaction using a customized 55-mer primer containing a 24-mer T7 promoter and random oligomer sequences (T7dT-N; SIGMA-Aldrich Japan, Komae, Japan). This was followed by an in vitro transcription reaction to incorporate amino allyl nucleotides into aRNA. An aliquot (10 μg) of synthesized aRNA was coupled with Cy3 or Cy5 dyes and purified on an Amicon YM-30 column (Millipore, Billerica, MA). Probes were hybridized to the DNA microarray at 57°C for 16 h, then excess probe was removed by washing with washing solution I (0.2 × SSC, 0.1% SDS) at 50°C for 10 min and then with washing solution II (0.05 × SSC) at room temperature for 5 min.
Signals on the DNA microarray were detected with a laser scanner GMS428 (Affymetrix, Santa Clara, CA) and signal intensities were calculated using ImaGene ver. 6.0 (BioDiscovery, El Segundo, CA). Signal intensity of each spot was calculated automatically with the subtraction of local background value. Each signal value was normalized against the averaged intensity of internal control spots. The microarray data of cycloheximide assay were further normalized due to relatively high local background intensity that causes a number of false-positive errors especially in low-intensity signal data. Thus, we manually performed careful local background correction in each spot and flooring treatment by adding a constant value to all data of signal intensity due to Cy3 and Cy5 signals. The control experiment with wild-type cells (Supplementary Fig. S1) demonstrated that the range of experimental errors in the induction factor was between 0.5 and 2.0, as indicated by dashed lines in Supplementary Figs S1 and S2. Clustering analysis of data was carried out using MeV software (TIGR; http://www.tm4.org/mev.html).
2.4. Western analysis of sigma factors
To clone genes encoding the C. merolae sigma factors SIG1, SIG2, SIG3, and SIG4, each open reading frame was amplified by PCR using C. merolae genomic DNA as the template. Each PCR-amplified fragment was inserted into pENTR™/D-TOPO (Invitrogen) according to the manufacturer's instructions. To prepare the antigen for production of polyclonal antibodies, each cloned gene was recombined into the destination vector pDEST-HIS.30 Preparation and purification of polyclonal antibodies and immunoblot analyses were carried out as described previously.30,31
3. Results
3.1. Design of microarray for analysis of organelle gene expression in C. merolae
The eukaryotic red alga C. merolae has a fully sequenced genome, and is an important model organism for studying the evolution of primitive photosynthetic eukaryotes.9 In this study, we monitored organelle gene expression in C. merolae during synchronous cultivation to understand the relationship between organelle gene expression and cell cycle events, and to determine whether gene expression in the chloroplast was correlated with that in the mitochondrion. Previously, we prepared and analysed a custom DNA microarray focused on chloroplast genes.26 Here, we prepared a new custom DNA microarray covering both the chloroplast and mitochondrial genomes. This enabled comparative analysis of gene expression in both organelles. We designed 50-mer oligonucleotide probes from a highly specific portion of each ORF of the chloroplast and mitochondrial genomes (Supplementary Table SI). These probes were spotted onto a glass slide as a duplicate for statistical reproduction. The results of a control experiment confirmed that experimental errors of our microarray system were low (within a 2-fold difference; Supplementary Fig. S1).
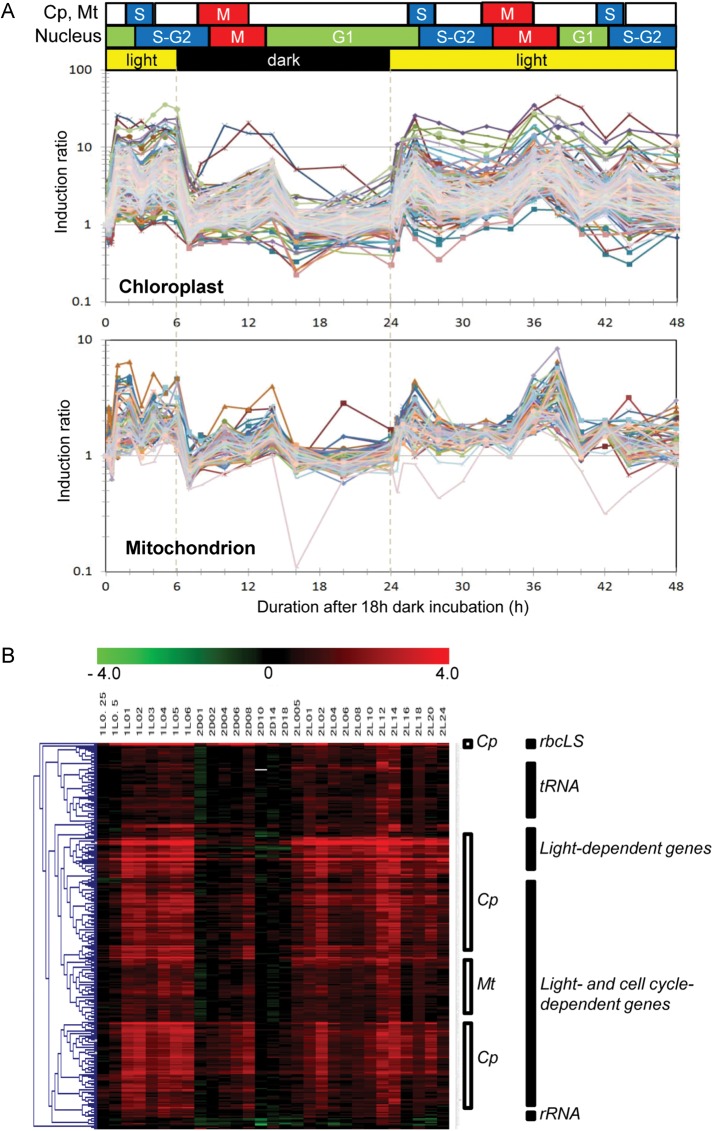
Several groups have established synchronous culture systems for C. merolae.5,13 We reconstructed a synchronous culture system with some modifications that optimized cell growth in the MA2 medium.28 Cultures (5 L) were grown in a thermostat jar fermenter and were bubbled with 2% CO2 in air and illuminated with white light (150 μE m−2 s−1). Cells were first incubated for 18 h in the dark, and then incubated for 6 h in the light before incubation for a further 18 h in the dark. Then, cells were cultured in continuous light conditions for 24 h. The cell sample obtained after the first 18-h dark incubation (1D18) was assigned as the time 0 point of the synchronization culture (Fig. 1). Cell samples were collected at 28 time points during the 48-h incubation period (see upper part of Fig. 1B and legend of Supplementary Fig. S2). Sample names in Fig. 1B, such as 1L0.25 (0.25 h after first light illumination), 2D18 (18 h after onset of second dark period) or 2L24 (24 h after second light illumination) correspond to 0.25, 24, and 48 h time points, respectively, in Fig. 1A. The cellular mitotic indexes during incubation in a light–dark cycle and under constant light are shown in Supplementary Fig. S2.
Figure 1.
Expression profiles of organelle genes in DL and LL conditions. (A) The index of organelle division cycle (see also Supplementary Fig. S2) and expression profiles of 243 genes in the plastid genome and 62 genes in the mitochondrial genome are shown. The horizontal axis shows the duration of incubation from the onset of light illumination at starting time point of synchronization culture (time 0). The vertical axis shows the induction ratio of each gene compared with the basal level of expression at time 0. Expression profiles of chloroplast genes (upper panel) and mitochondrial genes (lower panel) were highly synchronized. (B) Hierarchical tree view of plastid and mitochondrial genes. Chloroplast genes (Cp), mitochondrial genes (Mt), and other functional categories are indicated. Each column of the heat map corresponds to each time point of sampling for DNA microarray analysis. Log2 values of the induction ratio of each gene compared with the basal level of expression at time 0 are shown in red (induced genes) or green (suppressed genes; indicator is shown at the top of the figure). Sample names in (B), such as 1L0.25 (0.25 h after first light illumination), 2D18 (18 h after the onset of second dark period) or 2L24 (24 h after second light illumination) correspond to 0.25, 24, and 48 h time points from time 0 in (A), respectively. Details of cellular mitotic indexes during incubation in a light–dark cycle and under constant light are shown in Supplementary Fig. S2. Functionally related genes were categorized into the same sub-clusters. The list of DNA microarray data is available at the following web site (http://merolae.biol.s.u-tokyo.ac.jp/download/).
3.2. Coordination of the accumulation of organelle transcripts with the cell cycle phase
Previously, a genome-wide analysis reported that steady-state transcript levels in mitochondria do not vary between light and dark phases and are stable throughout the diurnal time course in Arabidopsis thaliana.32 Another DNA microarray analysis in Nicotiana tabacum also showed that transcript levels of about half of plastid genes were not changed during light and dark conditions.33 Northern analyses in Physcomitrella patens and in C. reinhardtii, several chloroplast genes showed almost constitutive expression in diurnal conditions.34,35 However, it is not clear that these organisms do not have synchronous mechanisms to regulate the transcript levels of organelle genes in a genome-wide manner or this synchrony is difficult to detect by perturbation due to averaged results of numerous organelles in each cell in individual phases of cell cycle. Since C. merolae has only one mitochondrion and one chloroplast, we anticipated that changes in organelle transcript levels in response to external and internal conditions may be more easily recognizable in this organism. Thus, we analysed the expression profiles of genes of C. merolae organelle genomes during synchronous cultivation (Fig. 1A). As expected, expressions of chloroplast and mitochondrial genes were strongly induced by light. We also found that a number of gene transcripts of both organelles showed transient accumulation around 4–8 h after the onset of the dark period. This accumulation also occurred with similar timing during continuous light illumination, which corresponded to 36–38 h after the onset of the first light illumination (Fig. 1A). Thus, this expression pattern appeared to reflect responses to internal events, rather than environmental light conditions. For both organelles, the peaks in transcript accumulation occurred at similar times. This indicated that the accumulation of transcripts from both organelles is coordinately regulated. Microscopic observation revealed that cells were most commonly at the M-G1 phases at these time points (see also Supplementary Fig. S2). Thus, the transient accumulations of most organelle transcripts were due to cell cycle-dependent regulation.
The peak height of the mitotic index curve gradually diminished and finally became asynchronous due to the diffusion of cell cycle phases among the cells in the culture (Supplementary Fig. S2). Similarly, expression patterns of organelle genes also partly diffused after 42 h from the start of incubation (time 0; Fig. 1). These results provide further evidence that the organelle gene expression pattern is highly correlated with cell cycle events rather than circadian control. In another study, qPCR analysis of the expressions of 74 selected genes (70 nuclear genes and 4 plastid genes) during light–dark cycles14 showed that the expression of a group of light-inducible genes, e.g. the nuclear-encoded atp1 gene for the alpha subunit of mitochondrial ATPase, was transiently induced after M phase during a dark period. Fujiwara et al.12 also detected the accumulation of several transcripts of nuclear-encoded organelle genes, which were specific to S phase or to G2 phase, during a dark period. These two reports indicated that phases of the nuclear cell cycle might affect organelle gene expression and metabolism. Together with our data, these results suggest that there is strong coordination of gene expression among the nucleus, the chloroplast, and the mitochondrion in a cell cycle-dependent manner.
3.3. Highly coordinated gene expression of chloroplast and mitochondrial genomes
The expression patterns of chloroplast and mitochondrial genes were very similar (Fig. 1). To estimate the similarity of gene expression patterns of the two organelles by a statistical method, we performed a relative network analysis by calculating the correlation coefficient of the expression profile of each gene using the RelNet function of MeV software (http://www.tm4.org/mev.html; Supplementary Fig. S3). The matrix of the calculated correlation coefficient (R2) for all genes and the list of DNA microarray data are available at the following web site (http://merolae.biol.s.u-tokyo.ac.jp/download/). The results of correlation clustering showed that the co-expression network, R2 > 0.7, covers 302 genes (98%) out of a total of 306 genes in the organelle genomes. This large network included almost all genes of organelle genomes. Expression profiles of only four genes, such as two rRNA genes (CMV029R, CMV032R) and two mitochondrial genes, CMW051R (trnS) and CMW002C (sdhB), were not categorized into this large group. Unfortunately, we could not identify oscillatory patterns from these four expression profiles.
3.4. Highly similar expression patterns of functionally related genes
In this study, we observed that genes in the same functional category showed very similar expression patterns. In addition, there were only minor differences in gene expression patterns between the chloroplast and the mitochondrion. To clearly illustrate the results of the correlation analyses, we constructed a tree view of hierarchical clustering according to Pearson correlation coefficients (Fig. 1B). When expression profiles of genes were compared, genes that were functionally related showed much higher correlation coefficients than those of unrelated genes. Thus, functionally related genes assembled into several sub-clusters in the hierarchical tree view. For instance, the tRNA cluster contains 51 genes including 20 tRNAs of both organelles. The light-dependent gene cluster contains 27 plastid genes of photosynthetic machineries. The light- and cell cycle-dependent cluster contains >200 genes of both organelles. The housekeeping gene cluster contains 10 genes such as rRNAs, RNaseP and tmRNA. The genes in the same sub-clusters were not from the same operon or gene cluster in the organelle genomes, except for a large gene cluster of the ribosomal proteins and ATP synthase subunits.7 This result indicates that there is coordinated expression of functionally related genes, although the mechanism by which this coordination occurs is unknown. Mitochondrial and chloroplast genes were also roughly divided into distinct sub-clusters as shown in Fig. 1B.
3.5. Classification of gene expression patterns entrained by light or cell cycle phases
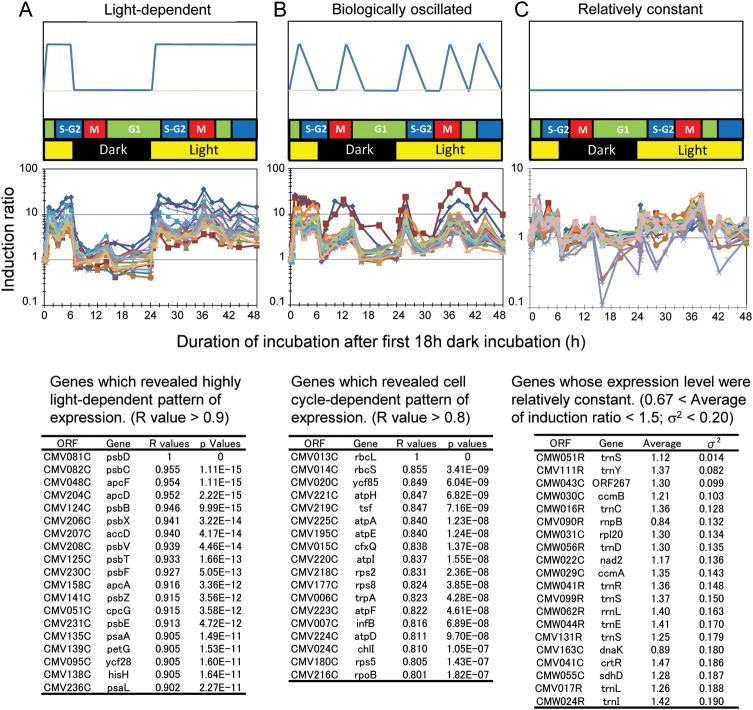
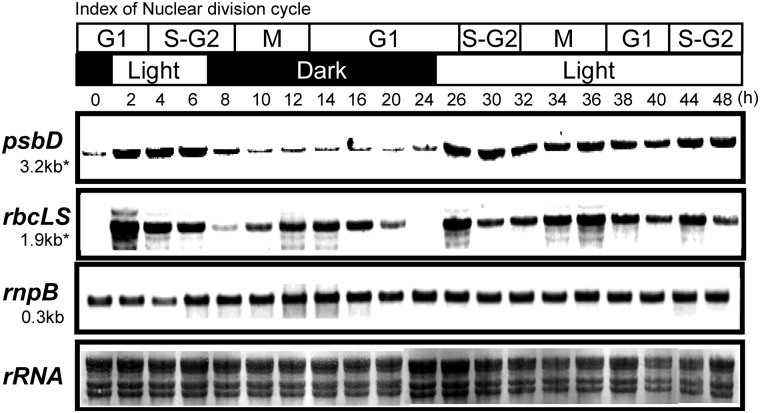
Next, we used a template-matching approach36,37 to identify genes whose expressions were highly responsive to light or internal phases (Fig. 2). Genes were grouped by their correlation coefficients (i.e. the similarity of their expression to those of the template genes). According to the results shown in Fig. 1, we chose psbD and rbcL genes as template expression profiles for typical light-responsive and cell cycle-responsive genes, respectively, and confirmed their expression patterns by northern analysis (Fig. 3). Using the PTM calculation function of MeV software, we determined the correlation between the expression pattern of each gene and those of the two template genes. That is, we determined whether and to what extent the expression of each gene resembled that of psbD or rbcL. These correlations are expressed as R2 values (Fig. 2). The psbD-correlated group (Fig. 2A) included many genes for photosynthetic machineries, and their expression patterns were highly light dependent. Some of these genes consisted of operons, but most were located at distinct loci on the chloroplast genome. The expressions of the genes in this group were slightly induced at the end of the second dark period (Fig. 2A), which is likely due to circadian oscillation. The rbcL-correlated group (Fig. 2B) included genes for translation machineries, ATP synthase, and so on. The expression of these genes showed transient induction just after the onset of light illumination as well as at the M-G1 phase (Fig. 2B). Some genes were expressed at relatively constant levels throughout the experimental period (Fig. 2C). This group (Fig. 2C) contained a number of tRNAs and several housekeeping genes, such as the rnpB gene. Even in this group, gene expression was increased or decreased at several time points during the cell cycle. The expression of the rnpB gene in Cluster III was confirmed by northern analysis (Fig. 3).
Figure 2.
Template-matching analysis to identify light-dependent, cell cycle-dependent, or relatively constant genes in chloroplast and mitochondrial genomes. (A) Light-dependent genes showed a high correlation coefficient with the psbD gene, whose expression is highly light specific. (B) Cell cycle-dependent genes showed a high correlation coefficient with the rbcL gene, whose expression is highly specific to cell cycle phases. (C) Genes expressed at relatively constant levels. Upper panels show model pattern of gene expression used as a template for the calculation of the correlation coefficient in each group. A typical gene, which was calculated as highly similar pattern with that of the template pattern, was used as a second template. Lower panels show the list of genes and their respective values of the correlation coefficient with the expression pattern of the template genes. Middle panels show expression profiles of the genes, which are shown in the lower panels. The matrix of the calculated correlation coefficient (R2) for all genes is available at the following web site (http://merolae.biol.s.u-tokyo.ac.jp/download/).
Figure 3.
Northern analysis of typical genes in each cluster in Fig. 2. The psbD and rbsLS transcripts were accumulated in a light-dependent manner or in cell cycle-dependent manner, respectively. Cells were collected at indicated time points during synchronous cultivation as same as Fig. 1. The preparation of the probes of psbD, rbcL, and rnpB genes was described in Section 2. A methylene blue-stained membrane was shown in the bottom panel to show the amount of rRNAs.
Next, we searched for conserved elements in the 5′ upstream and 3′ downstream regions of the genes in each group using the web-based tool Melina II (http://melina2.hgc.jp/public/index.html). No conserved elements were found in the 5′ upstream region; however, we identified a conserved element in the 3′ downstream region of genes in each group by the program: CONSENSUS and MD scan in the Melina II [Supplementary Fig. S5 (element found in the light-inducible genes) and 6 (element found in the cell cycle-dependent genes)]. These elements were found in most genes in each group. The ORFs, which does not possess this element in their 3′ downstream region was not the terminal gene of the putative operon. Positions of each element in the 120 base of 3′ downstream region of each gene were shown in Supplementary Figs S5 and S6. Several genes have plural number of the elements in their 3′ downstream region.
3.6. Expression of nuclear-encoded sigma factors of chloroplast RNA polymerase
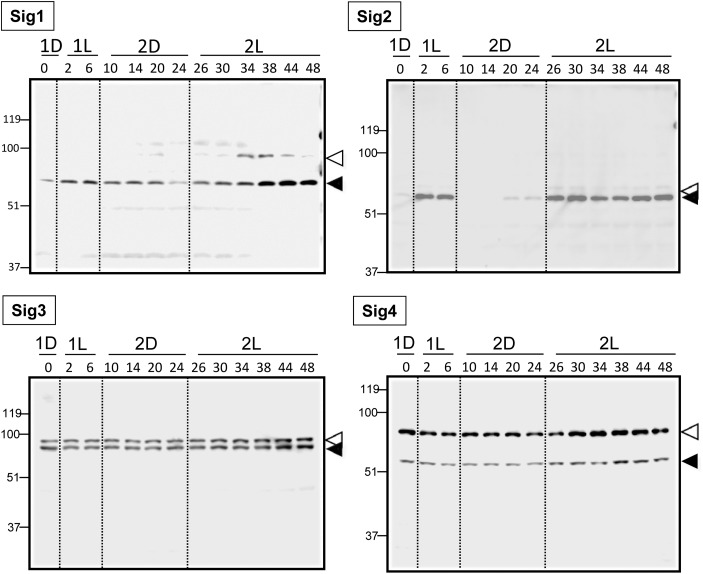
As described in Section 1, PEP appears to be the sole chloroplast transcription machinery in C. merolae. Four nuclear genes, CMK044C, CMQ213C, CMR165C, and CMM072C are predicted to encode sigma factors for PEP, SIG1, SIG2, SIG3, and SIG4, respectively.26 Previously, it was shown that light induces the accumulation of transcripts of these four sigma factor genes.26 Thus, they are likely responsible for the light activation of chloroplast gene expression. To understand the link between chloroplast transcription and the expression of sigma factors, we analysed the expression of these four sigma factors at the protein level. First, we prepared four types of recombinant proteins (SIG1, SIG2, SIG3, and SIG4) using an Escherichia coli system, and then obtained specific antibodies against each protein (see Supplementary Fig. S4). Then, expression levels of the four sigma factors were determined by immunoblot analysis during synchronized cultivation in the same conditions as those shown in Fig. 1 (Fig. 4). In each case, we detected two bands, presumably the precursor and the processed mature protein. These bands were not detected in the corresponding preimmune sera (Supplementary Fig. S4). The size of each SIG precursor was predicted from the C. merolae genome database (http://merolae.biol.s.u-tokyo.ac.jp/), as follows: Sig1, 80 kDa; Sig2, 52 kDa; Sig3, 73 kDa; and Sig4, 67 kDa. Band sizes of CmSIG3 and CmSIG4 proteins were higher than that predicted from the database. It means some modifications such as phosphorylation38 of sigma factors might be occurring in vivo. In the case of SIG3 and SIG4, the band intensity of the precursor was much higher than that of the mature protein at several time points. This result suggests that sigma factor proteins may be regulated at the translocation and/or maturation steps. The amount of SIG1 protein increased after the onset of light illumination and also at specific cell cycle phases (M-G1 and G1-S), indicating that SIG1 may be involved in cell-cycle specific transcriptional activation. SIG2 protein was detected only in light conditions, suggesting that this sigma factor may be involved in light-dependent chloroplast gene expression, which is observed for genes such as psbD (Fig. 3). SIG3 and SIG4 proteins were found constitutively during the dark–light (DL) and constant light (LL) cycles, although all four SIG gene expressions have been reported as light inducible.26 Therefore, expressions of SIG3 and SIG4 are probably regulated at translational or post-translational levels. These sigma factor proteins may contribute to the basal expression of housekeeping genes in the chloroplast. These results indicated that the expression of sigma factor proteins can categorized into three groups: cell cycle phase-specific, light-dependent, or relatively constant. These three groups roughly correlate with the results of the clustering analysis of the DNA microarray.
Figure 4.
The expression of proteins of nuclear-encoded sigma factors for plastid RNA polymerase. Cells were collected at indicated time points during synchronized culture. Protein extraction and western analyses were conducted as described in Section 2. Precursors and mature proteins are indicated by white and black triangles, respectively. Time point 1D18 corresponds to time 0 point in Fig. 1. The preparation and specificity of the antibodies were shown in Supplementary Fig. S4. The Sig2 protein showed a typical light-dependent accumulation.
3.7. Light-inducible organelle gene expression was inhibited by cycloheximide and dibromothymoquinone
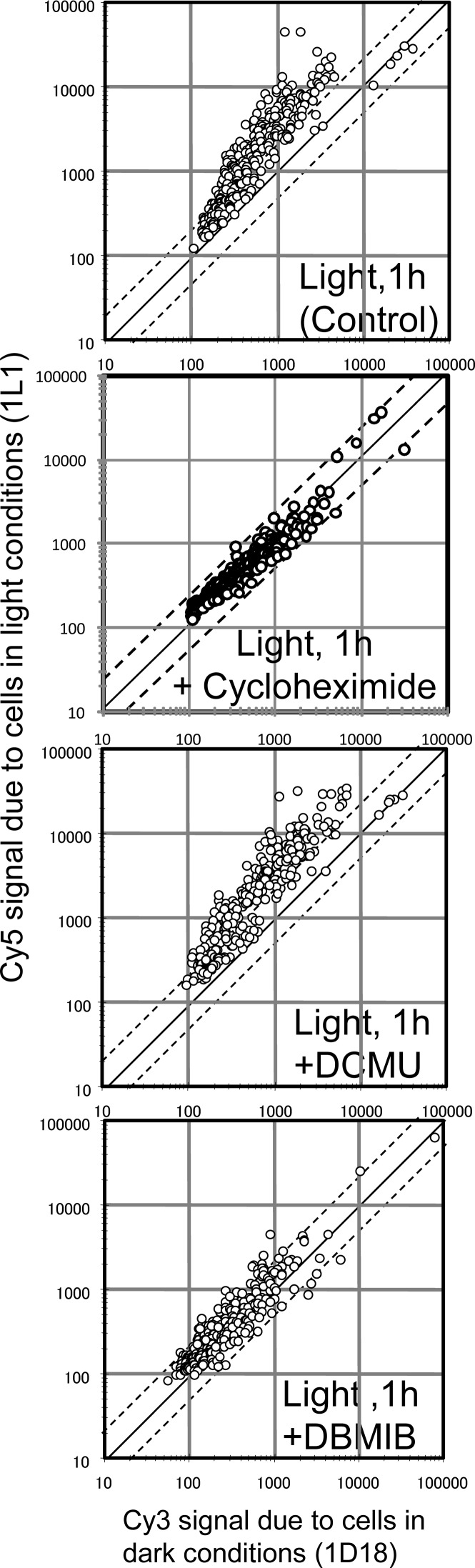
Next, we examined the signaling pathways involved in light-inducible gene expression in organelles using three inhibitors: cycloheximide, 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU), and dibromothymoquinone (DBMIB) (Fig. 5). Cells were treated with these inhibitors at the end of 18 h dark incubation (time 0 in Fig. 1 and Supplementary Fig. S2), and then subjected to light conditions for 1 h. In an experiment using the cytoplasmic translation inhibitor cycloheximide, Kawazoe et al.39 found that light-inducible plastid gene expression in Chlamydomonas required de novo protein synthesis in the cytoplasm. In C. merolae, de novo synthesis of cytoplasmic protein was also required for light-inducible organelle gene expression. Taking the results shown in Figs 4 and 5 into consideration, the inhibition of SIG1 and SIG2 protein synthesis may be responsible for the inhibitory effect of cycloheximide. We also determined the effects of the photosynthesis inhibitors DCMU and DBMIB. DCMU inhibits the electron transfer of QA to QB in the PSII complex, inhibiting linear electron flow.40 DCMU did not affect the accumulation of transcripts after the shift from dark to light. However, DBMIB, which is assumed to inhibit the access of reduced plastoquinone to the cytochrome b6f complex (Cyt b6f),41 causing the inhibition of both linear and cyclic electron flow, severely inhibited light-induced gene expression in both organelles.
Figure 5.
Effects of inhibitors on light-inducible gene expression in organelles. Inhibitors were added to cell cultures in dark conditions (1D18; time 0). Cells were then incubated for 1 h in light conditions (150 μEm−2s−1). Total RNA was extracted from cells and used for DNA microarray analyses. Experiments were reproduced twice in biologically independent conditions and yielded essentially the same results. Final concentrations of inhibitors were as follows: cycloheximide (100 μM), DCMU (10 μM), and DBMIB (5 μM). Suitable concentrations of DCMU and DBMIB that do not affect the respiration but inhibit the cellular proliferation, cell division and light-induced oxygen evolution were determined by oxygen electrode and microscopic analysis. Cycloheximide and DBMIB inhibited the light-inducible gene expression of organelle genomes.
3.8. Stability of chloroplast transcripts under light and dark conditions
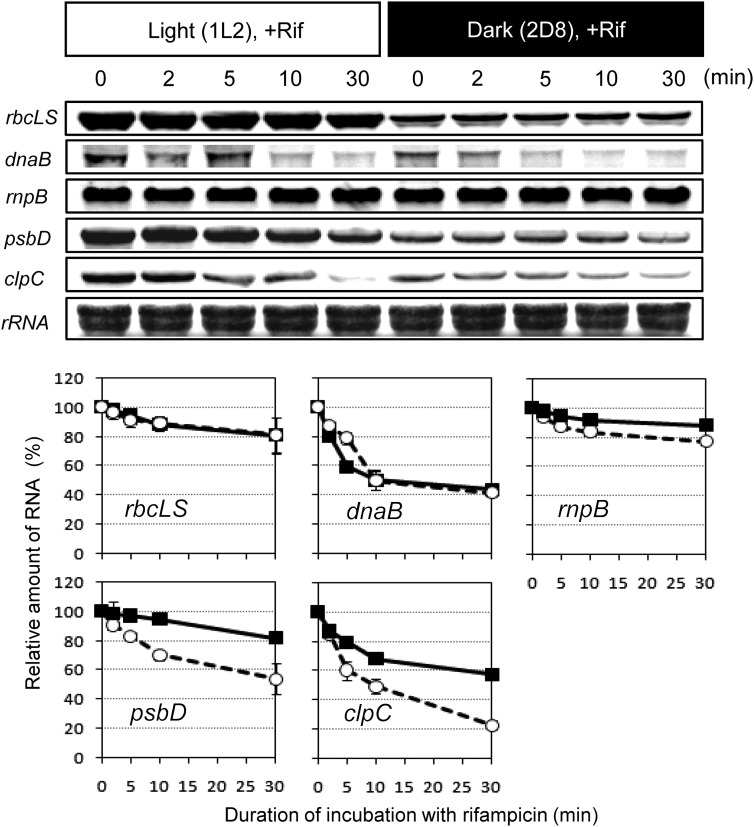
In vascular plants, accumulation of chloroplast transcripts is usually regulated at a post-transcriptional level, such as stability, modification, editing, and translation (reviewed in).42 Here, we used the rifampicin assay to examine changes in stability of several transcripts under light and dark conditions. Rifampicin is a specific inhibitor of the bacterial-type chloroplast RNA polymerase, and has been shown to be effective in red algae.43,44 Rifampicin was added to the cells 2 h after the first onset of light illumination (1L2) or 8 h after the second dark shift (2D8). As shown in Fig. 6, psbD and clpC transcripts were more stable during the light phase, suggesting regulation at the level of mRNA stability. However, the stability of rbcLS gene transcripts remained constant in light and dark conditions. Thus, the transient accumulation of rbcLS transcripts in the M-G1 phase (Fig. 3) during the dark period was likely due to transcriptional activation. Stability of rnpB transcripts, which showed a relatively constant expression pattern (Fig. 3), was also unaffected by the light conditions.
Figure 6.
Analysis of the stability of some mRNAs in the chloroplast. Rifampicin (100 μg/ml) was added at indicated time point and cells were collected before (time 0) and after treatment (2, 5, 10, and 30 min). Signal intensity shown in lower panels was normalized against rRNA. Values are means of three independent experiments ± standard deviation. The preparation of the probes of each gene was described in Section 2. A methylene blue-stained membrane was shown in the bottom panel to show the amount of rRNAs.
4. Discussion
4.1. Correlation between organelle gene expression and nuclear cell cycle phases
In this study, we examined profiles of chloroplast and mitochondrial gene expression in C. merolae and found that gene expression in both organelles was highly responsive to cell cycle control and environmental light conditions. As shown in Fig. 2, genes for transcription, translation, ATP synthesis, and RuBisCO are candidate genes whose expressions are obviously controlled by the cell cycle. However, it remains unclear how and why many organelle transcripts accumulated at the M-G1 phase, even in dark conditions. Cyanidioschyzon merolae is a photoautotrophic organism containing only one mitochondrion and one plastid, and a synchronous culture system entrained by light and dark cycles has been established. Thus, C. merolae is an ideal system for studying cell cycle-coordinated gene expression in organelles, because unlike other organelle analysis systems, it does not have multiple organelles with asynchronous replication. In vascular and other green plants, it is very difficult to obtain synchrony of proliferation cycles because of the abundant mitochondria and chloroplasts in the cell. In a previous study with tobacco BY-2 culture cells, use of organelle DNA replication inhibitor, nalidixic acid, could synchronously arrest the organelle proliferation cycle, and thus we could successfully observe the organelle-to-nucleus signaling pathway and its regulatory mechanisms.4,45,46 This kind of approach may help the understanding of events occurring in cells containing multiple organelles.
4.2. Nuclear factors for light-inducible gene expression in the organelles
Inhibition of cytosolic translation by cycloheximide abolished the light-induced transcriptional activation of genes from both organelles. This result suggests the involvement of nuclear-encoded proteins in transcriptional activation (Fig. 5). As described in Section 1, the nuclear genome of C. merolae encodes only one NEP-type RNA polymerase that is predicted to be targeted into the mitochondrion, plastid transcription is likely dependent only on PEP. Since the PEP activity and the specificity are dominated by nuclear-encoded sigma subunits, it is reasonable to consider that the expression of sigma factors is responsible for the control of the plastid transcriptome. As shown in Fig. 4, accumulation profile of SIG1 and SIG2 proteins showed the cell-cycle correlated pattern and the light-dependent pattern, respectively. These patterns well correlate with the expression profiles of cell-cycle correlated genes and light-dependent genes in the plastid genome, respectively, as shown in Fig. 2. Consistently, inhibition of light-inducible de novo synthesis of these SIG proteins by cycloheximide abolished the light-inducible expression of plastid genes (Fig. 5). These results strongly suggest the sigma factor-dependent molecular mechanism for the correlations provided in this study. On the other hand, cycloheximide did not affect the basal levels of transcripts in the organelles (Fig. 5). Thus, it is likely that the basal accumulation of transcripts is dependent on the constitutively expressed sigma factors, SIG3 and/or SIG4. Light induction of mitochondrial gene expression could also be regulated by nuclear-encoded factors including the NEP-type mitochondrial RNA polymerase (CMJ257C) and other regulator(s). The effects of cycloheximide on mitochondrial gene expression (Fig. 5) may be explained by inhibition of these proteins synthesized de novo in the cytoplasm.
In a green alga, C. reinhardtii which has no NEP in chloroplasts and it seems to have only one sigma factor,47,48 the plastid gene expression is limited mainly to the post-transcriptional level.49,50 Thus, the sigma factor in C. reinhardtii is considered as just a factor for basal transcription. However, our results suggest the red alga, C. merolae, selectively uses four sigma factors depending on the cellular conditions similarly with vascular plants. These results must contribute to the research field of sigma factor-mediated regulation of plastid transcription.
4.3. Light-inducible organelle gene expression is affected by a DBMIB-related process
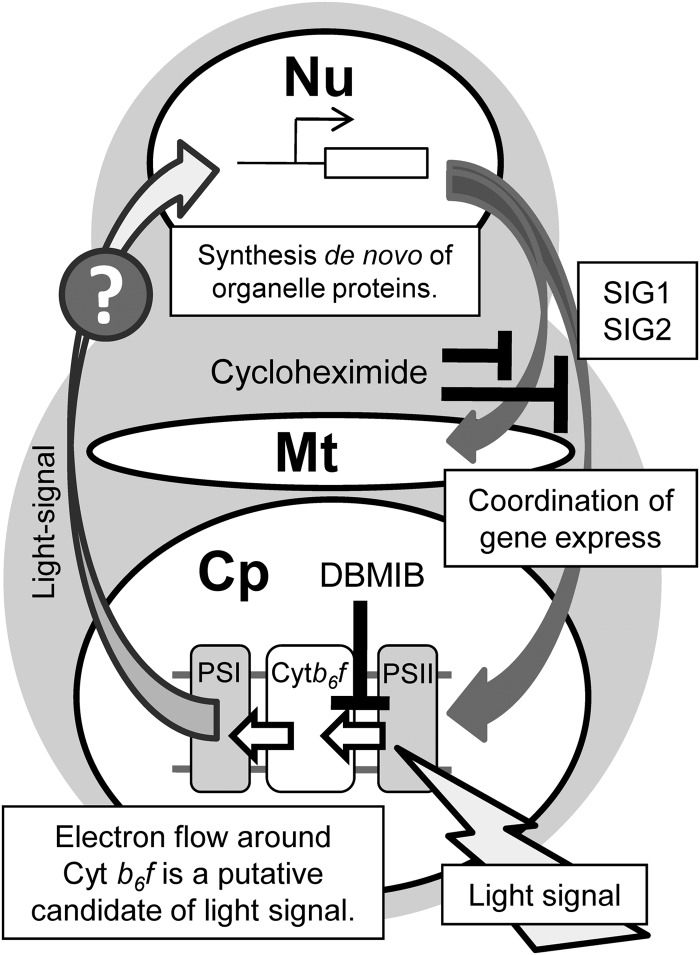
What senses the light for the light-inducible organelle gene expression in C. merolae? Our results revealed that a process inhibited by DBMIB is essential for light-inducible gene expression in both organelles. These results, together with the results of cycloheximide addition (Fig. 5) suggest that de novo synthesis of specific nuclear-encoded protein(s) is required. The expression of these proteins is dependent on light sensing around the thylakoid membrane and subsequent signal transduction to the nucleus. It has been argued that the photosynthesis-driven redox state is involved in light-dependent transcript accumulation in chloroplasts, i.e. the redox state is the mechanism for light-dependent regulation of chloroplast transcription. For example, it was suggested that the plastoquinol/plastoquinone ratio in the thylakoid membrane affects the expression of the chloroplast psbA and psaAB genes.51 On the other hand, Matsuo and Obokata52 concluded that induction of the expression of Chlamydomonas PSI mRNAs is dependent on light illumination, but not on the redox state of plastoquinone. Our results indicate that the light-inducible gene expression in chloroplast does not depend on the redox state of plastoquinone pool, but depending on the integrity of the plastoquinol-oxidizing site of the Cytb6f complex. A report also suggested that a Cyt b6f-related factor, which is inhibited by DBMIB, affects the light-induced expression of several nuclear-encoded plastid proteins in Chlamydomonas.53 In C. merolae, our results suggested that a Cyt b6f-related factor is crucial for sensing light and subsequent transcriptional activation (Fig. 7). Cyanidioschyzon merolae possesses neither phytochrome nor phototropin as photoreceptors, but has five nuclear genes that encode cryptochrome-related proteins. However, blue-light-dependent gene expression was not observed in C. merolae (unpublished observations), and thus, the photosynthetic electron transfer process in the thylakoid membrane may itself function as the light sensor for transcriptional activation. The inhibitory effect of DBMIB is consistent with this scheme, whereas the signaling process from the chloroplast to the nucleus remains unknown.
Figure 7.
Hypothetical scheme of coordinated gene expression in the plastid and mitochondrion. Nu, nucleus; Mt, mitochondrion; Cp, chloroplast.
4.4. Correlation between changes of chloroplast and mitochondrial transcriptomes
This study represents the first report of highly synchronous global gene expression profiles in the chloroplast and mitochondrion. This phenomenon is probably due to synchronous activation of nuclear-encoded factors, such as SIG proteins. It is likely that this synchrony is essential for coordination of the functions of these two organelles in light- and cell cycle-dependent manners. The successful functioning of important metabolic pathways including lipid biosynthesis, tetrapyrrole biosynthesis, and photorespiration rely on inter-organelle exchanges of metabolites. The coordinated activity of the two organelles would be important to optimize these reactions. In another study, impaired chloroplast development affected steady-state transcript levels in mitochondria in the barley albostrians mutant.54 Our synchronous system clearly showed that coordination of gene expression between the plastid and mitochondrion is dynamically modulated by the light and cell cycle in C. merolae.
4.5. Correlations among changes in transcript levels of functionally related genes
As in bacterial transcriptional regulation, organelle-encoded genes tend to be transcribed as poly-cistronic RNAs from a limited number of transcriptional start sites. It is well known, especially in chloroplasts of green lineages, that such large transcripts are subsequently processed into smaller units, and that gene expression is primarily regulated at the post-transcriptional level. In this study, we performed cluster analysis for C. merolae organelle transcripts, and found that chloroplast genes with the same function frequently show similar expression profiles, even when their chromosomal loci are not linked to each other (Fig. 2). These data indicated the presence of a common regulator and regulatory elements for each cluster. As discussed above, sigma factors for the chloroplast RNA polymerase may function as the transcriptional regulators for these clusters. Recently, Minoda et al.55 clarified that a plastid-encoded transcription factor, Ycf30 (CbbR), is able to bind to the promoter region of the rbcLS operon and up-regulates their expression in the isolated-chloroplasts of C. merolae. Together with three other transcription factors encoded in the plastid genome, these regulators may also be involved in the correlated gene expressions. In addition, we found conserved sequence elements within the 3′ regions of light-dependent (Fig. 2A) and cell cycle-dependent (Fig. 2B) genes. In Chlamydomonas, the endonuclease CSP41, which binds the stem-loop structure of the 3′ untranslated region of transcripts,56,57 is a major regulator of transcript stability. Two homologs of CSP41 in C. merolae, CMH047C and CML047C, are candidates for such post-transcriptional controls. PPR proteins encoded by the nuclear genome may also have roles in the modulation of mRNA processing and the stability.58
4.6. Effect of circadian rhythms on organelle gene expression
Expression of photosynthetic genes, such as the psbD gene in the chloroplast genome, and the nuclear-encoded SIG2 proteinshowed weak induction at the time point of artificial dawn, suggesting that gene expression showed a circadian rhythm (Figs. 3 and 4). In Chlamydomonas, nuclear regulation of chloroplast circadian period was observed.59 In C. merolae, it also seems that the factor due to the nuclear circadian rhythm affects the expression of SIG2 protein, and SIG2 entrain the expression pattern of the photosynthetic genes, such as the psbD with that of nuclear rhythms. In terms of cell-cycle control, the expression patterns of organelle genes appeared to be dominated by the index of cell division. These observations indicate that light and cell cycle controls are more important factors in the patterns of organelle gene expression than the circadian rhythm in C. merolae.
4.7. Fundamental differences between C. merolae and other organisms
In our system, synchrony of transcript levels of organelle genes was detected in C. merolae. In vascular plants, such coordination of organelle gene expression has not been observed so far. One possible reason is that organelle proliferation cycle and division cycles are not coordinated with the cell cycle in mature plant cells. Second possible reason is that they do not have any synchronous mechanism to regulate the transcript levels of organelle genes in a genome-wide manner. Third possibility is that the synchrony is difficult to detect due to averaged results from unsynchronized organelles in each cell throughout the cell cycle. To discriminate these three possibilities, it would be helpful to check the coordination of organelle gene expression during the specific experimental conditions where organelle proliferation occurs synchronously in plant cells. As Kobayashi et al.4 showed in tobacco BY-2 cells, organelle proliferation synchronously begins after transferring the stationary phased cells to new culture media. Using the similar experimental condition, some coordinated pattern of mitochondrial gene expression was observed by EST and microarray analyses.60 While only fragmental data have been obtained from vascular plants, this kind of evidence may suggest the presence of coordination mechanism for organelle transcriptome in vascular plants in specific conditions such as developing organ, where the mitochondrial DNA replication is highly activated.61 Available information suggests that BY-2 cells could be a good model system to investigate the relationship of organelle transcription and organelle proliferation. Recently, Maple et al.62 found that a nuclear-encoded gene whose transcript level was commonly affected in three arc mutants with altered plastid division phenotypes. This report indicates the relationship between plastid division cycle and cellular gene expression in plant cells. However, in case of mature cells of higher eukaryotic cells, mitochondria divide or fuse almost randomly and a part of them does not contain any DNA.63,64 Thus, the relationship of mitochondrial division cycle and cellular gene expression in plant cells is still unclear.
Why is not organelle proliferation in other species more synchronous? Chloroplasts and mitochondria are considered to have originated from bacterial symbionts and thus are possessing high autonomy as the nature of independent organism. Thus, it is not surprising that their proliferation and division cycles do not synchronize in a cell containing multiple organelles. On the other hand, the host cell and the endosymbionts must have evolved tight coordination mechanism to couple the organelle proliferation and division cycles with the cell cycle of the host cell, which should be most apparent when the number of organelle is only one in a cell. Details of these coordination mechanisms are still elusive, and to be clarified in future studies.
Supplementary Material
Acknowledgements
We thank Drs T. Fujiwara, O. Misumi, and T. Kuroiwa for technical discussion of microarray analysis of C. merolae.
Footnotes
Edited by: Katsumi lsono
Supplementary Data
Supplementary Data are available at www.dnaresearch.oxfordjournals.org.
Funding
This study was supported by Grants-in-Aid for Creative Scientific Research (16GS0304), for Scientific Research on Innovative Areas (23120505) and for Scientific Research (B; 21370015) to K.T. from Japan Society for Promotion of Science (JSPS), by a Grant-in-Aid for Scientific Research (20917006) to Y.K. from Ministry of Education, Culture, Sports, Science and Technology, Japan, and partly by the Sasakawa Scientific Research Grant from the Japan Science Society (19-455). S.I. is supported by a research fellowship from JSPS and a Grant-in-Aid for JSPS Fellows from the Ministry of Education, Science, and Culture. A.M. is supported by the Sasakawa Scientific Research Grant from the Japan Science Society.
References
- 1.Andersson S.G.E., Karlberg O., Canbäck B., Kurland C.G. On the origin of mitochondria: a genomics perspective. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2003;358:165–77. doi: 10.1098/rstb.2002.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gould S.B., Waller R.F., McFadden G.I. Plastid evolution. Annu. Rev. Plant Biol. 2008;59:491–517. doi: 10.1146/annurev.arplant.59.032607.092915. [DOI] [PubMed] [Google Scholar]
- 3.Kuroiwa T. The primitive red algae Cyanidium caldarium and Cyanidioschyzon merolae as model system for investigating the dividing apparatus of mitochondria and plastids. Bioessays. 1998;20:344–54. [Google Scholar]
- 4.Kobayashi Y., Kanesaki Y., Tanaka A., Kuroiwa H., Kuroiwa T., Tanaka K. Tetrapyrrole signal as a cell-cycle coordinator from organelle to nuclear DNA replication in plant cells. Proc. Natl Acad. Sci. USA. 2009;106:803–7. doi: 10.1073/pnas.0804270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyagishima S., Itoh R., Aita S., Kuroiwa H., Kuroiwa T. Isolation of dividing chloroplasts with intact plastid-dividing rings from a synchronous culture of the unicellular red alga Cyanidioschyzon merolae. Planta. 1999;209:371–5. doi: 10.1007/s004250050645. [DOI] [PubMed] [Google Scholar]
- 6.Ohta N., Sato N., Kuroiwa T. Structure and organization of the mitochondrial genome of the unicellular red alga Cyanidioschyzon merolae deduced from the complete nucleotide sequence. Nucleic Acids Res. 1998;26:5190–298. doi: 10.1093/nar/26.22.5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohta N., Matsuzaki M., Misumi O., et al. Complete sequence and analysis of the plastid genome of the unicellular red alga Cyanidioschyzon merolae. DNA Res. 2003;10:67–77. doi: 10.1093/dnares/10.2.67. [DOI] [PubMed] [Google Scholar]
- 8.Matsuzaki M., Misumi O., Shin-I T., et al. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature. 2004;428:653–7. doi: 10.1038/nature02398. [DOI] [PubMed] [Google Scholar]
- 9.Nozaki H., Takano H., Misumi O., et al. A 100%-complete sequence reveals unusually simple genomic features in the hot-spring red alga Cyanidioschyzon merolae. BMC Biol. 2007;5:28. doi: 10.1186/1741-7007-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nozaki H., Matsuzaki M., Takahara M., et al. The phylogenetic position of red algae revealed by multiple nuclear genes from mitochondria-containing eukaryotes and an alternative hypothesis on the origin of plastids. J. Mol. Evol. 2003;56:485–97. doi: 10.1007/s00239-002-2419-9. [DOI] [PubMed] [Google Scholar]
- 11.Ball S.G. Eukaryotic microalgae genomics. The essence of being a plant. Plant Physiol. 2005;137:397–8. doi: 10.1104/pp.104.900136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujiwara T., Misumi O., Tashiro K., et al. Periodic gene expression patterns during the highly synchronized cell nucleus and organelle division cycles in the unicellular red alga Cyanidioschyzon merolae. DNA Res. 2009;16:59–72. doi: 10.1093/dnares/dsn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moriyama T., Terasawa K., Fujiwara M., Sato N. Purification and characterization of organellar DNA polymerases in the red alga Cyanidioschyzon merolae. FEBS J. 2008;275:2899–918. doi: 10.1111/j.1742-4658.2008.06426.x. [DOI] [PubMed] [Google Scholar]
- 14.Moriyama T., Terasawa K., Sekine K., et al. Characterization of cell-cycle-driven and light-driven gene expression in a synchronous culture system in the unicellular rhodophyte Cyanidioschyzon merolae. Microbiology. 2010;156:1730–7. doi: 10.1099/mic.0.037754-0. [DOI] [PubMed] [Google Scholar]
- 15.Sugiura M. The chloroplast genome. Plant Mol. Biol. 1992;19:149–68. doi: 10.1007/BF00015612. [DOI] [PubMed] [Google Scholar]
- 16.Wakasugi T., Tsudzuki T., Sugiura M. The genomics of land plant chloroplasts: gene content and alteration of genomic information by RNA editing. Photosynth. Res. 2001;70:107–18. doi: 10.1023/A:1013892009589. [DOI] [PubMed] [Google Scholar]
- 17.Hajdukiewicz P.T., Allison L.A., Maliga P. The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J. 1997;16:4041–8. doi: 10.1093/emboj/16.13.4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hedtke B., Börner T., Weihe A. Mitochondrial and chloroplast phage-type RNA polymerases in Arabidopsis. Science. 1997;277:809–11. doi: 10.1126/science.277.5327.809. [DOI] [PubMed] [Google Scholar]
- 19.Silhavy D., Maliga P. Mapping of promoters for the nucleus-encoded plastid RNA polymerase (NEP) in the iojap maize mutant. Curr. Genet. 1998;33:340–4. doi: 10.1007/s002940050345. [DOI] [PubMed] [Google Scholar]
- 20.Cahoon A.B., Harris F.M., Stern D.B. Analysis of developing maize plastids reveals two mRNA stability classes correlating with RNA polymerase type. EMBO Repo. 2004;5:801–6. doi: 10.1038/sj.embor.7400202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sugita M., Sugiura M. Regulation of gene expression in chloroplasts of higher plants. Plant Mol. Biol. 1996;32:315–26. doi: 10.1007/BF00039388. [DOI] [PubMed] [Google Scholar]
- 22.Eibl C., Zou Z., Beck A., Kim M., Mullet J., Koop H.U. In vivo analysis of plastid psbA, rbcL and rpl32 UTR elements by chloroplast transformation: tobacco plastid gene expression is controlled by modulation of transcript levels and translation efficiency. Plant J. 1999;19:333–45. doi: 10.1046/j.1365-313x.1999.00543.x. [DOI] [PubMed] [Google Scholar]
- 23.Barkan A., Goldschmidt-Clermont M. Participation of nuclear genes in chloroplast gene expression. Biochimie. 2000;82:559–72. doi: 10.1016/s0300-9084(00)00602-7. [DOI] [PubMed] [Google Scholar]
- 24.Nickelsen J. Chloroplast RNA-binding proteins. Curr. Genet. 2003;43:392–9. doi: 10.1007/s00294-003-0425-0. [DOI] [PubMed] [Google Scholar]
- 25.Hess W.R., Börner T. Organellar RNA polymerases of higher plants. Int. Rev. Cytol. 1999;190:1–59. doi: 10.1016/s0074-7696(08)62145-2. [DOI] [PubMed] [Google Scholar]
- 26.Minoda A., Nagasawa K., Hanaoka M., Horiuchi M., Takahashi H., Tanaka K. Microarray profiling of plastid gene expression in a unicellular red alga, Cyanidioschyzon merolae. Plant Mol. Biol. 2005;59:375–85. doi: 10.1007/s11103-005-0182-1. [DOI] [PubMed] [Google Scholar]
- 27.Itoh R., Takahashi H., Toda K., Kuroiwa H., Kuroiwa T. Aphidicolin uncouples the chloroplast division cycle from the mitotic cycle in the unicellular red alga Cyanidioschyzon merolae. Eur. J. Cell Biol. 1996;71:303–10. [PubMed] [Google Scholar]
- 28.Ohnuma M., Yokoyama T., Inouye T., Sekine Y., Tanaka K. Polyethylene glycol (PEG)-mediated transient gene expression in a red alga, Cyanidioschyzon merolae 10D. Plant Cell Physiol. 2008;49:117–20. doi: 10.1093/pcp/pcm157. [DOI] [PubMed] [Google Scholar]
- 29.Minoda A., Sakagami R., Yagisawa F., Kuroiwa T., Tanaka K. Improvement of culture conditions and evidence for nuclear transformation by homologous recombination in a red alga, Cyanidioschyzon merolae 10D. Plant Cell Physiol. 2004;45:667–71. doi: 10.1093/pcp/pch087. [DOI] [PubMed] [Google Scholar]
- 30.Imamura S., Hanaoka M., Tanaka K. The plant-specific TFIIB-related protein, pBrp, is a general transcription factor for RNA polymerase I. EMBO J. 2008;27:2317–27. doi: 10.1038/emboj.2008.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Imamura S., Yoshihara S., Nakano S., et al. Role of sigma factors in controlling global gene expression in light/dark transitions in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Mol. Biol. 2003;325:857–72. [Google Scholar]
- 32.Okada S., Brennicke A. Transcript levels in plant mitochondria show a tight homeostasis during day and night. Mol. Genet. Genomics. 2006;276:71–8. doi: 10.1007/s00438-006-0119-7. [DOI] [PubMed] [Google Scholar]
- 33.Nakamura T., Furuhashi Y., Hasegawa K., et al. Array-based analysis on tobacco plastid transcripts: preparation of a genomic microarray containing all genes and all intergenic regions. Plant Cell Physiol. 2003;44:861–7. doi: 10.1093/pcp/pcg101. [DOI] [PubMed] [Google Scholar]
- 34.Ichikawa K., Shimizu A., Okada R., Satbhai S.B., Aoki S. The plastid sigma factor SIG5 is involved in the diurnal regulation of the chloroplast gene psbD in the moss Physcomitrella patens. FEBS Lett. 2008;582:405–9. doi: 10.1016/j.febslet.2007.12.034. [DOI] [PubMed] [Google Scholar]
- 35.Salvador M.L., Klein U., Bogorad L. Light-regulated and endogenous fluctuations of chloroplast transcript levels in Chlamydomonas. Regulation by transcription and RNA degradation. Plant J. 1993;3:213–9. doi: 10.1046/j.1365-313x.1993.t01-13-00999.x. [DOI] [PubMed] [Google Scholar]
- 36.Pavlidis P., Noble W.S. Analysis of strain and regional variation in gene expression in mouse brain. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-10-research0042. research0042.1–0042.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hulshizer R., Blalock E.M. Post hoc pattern matching: assigning significance to statistically defined expression patterns in single channel microarray data. BMC Bioinformatics. 2007;8:240. doi: 10.1186/1471-2105-8-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shimizu M., Kato H., Ogawa T., Kurachi A., Nakagawa Y., Kobayashi H. Sigma factor phosphorylation in the photosynthetic control of photosystem stoichiometry. Proc. Natl Acad. Sci. USA. 2010;107:10760–4. doi: 10.1073/pnas.0911692107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kawazoe R., Hwang S., Herrin D.L. Transcription of tufA and other chloroplast-encoded genes is controlled by a circadian clock in Chlamydomonas. Plant Mol. Biol. 2000;44:699–709. doi: 10.1023/a:1026519718992. [DOI] [PubMed] [Google Scholar]
- 40.Worland S.T., Yamagishi A., Isaacs S., Sauer K., Hearst J.E. Labeling quinone-binding sites in photosynthetic reaction centers: a 38-kilodalton protein associated with the acceptor side of photosystem II. Proc. Natl Acad. Sci. USA. 1987;84:1774–8. doi: 10.1073/pnas.84.7.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roberts A.G., Bowman M.K., Kramer D.M. The inhibitor DBMIB provides insight into the functional architecture of the Qo site in the cytochrome b6f complex. Biochemistry. 2004;43:7707–16. doi: 10.1021/bi049521f. [DOI] [PubMed] [Google Scholar]
- 42.Toyoshima T., Onda Y., Shiina T., Nakahira Y. Plastid transcription in higher plants. Crit. Rev. Plant Sci. 2005;24:59–81. [Google Scholar]
- 43.Troxler R.F., Lin S., Offner G.D. Heme regulates expression of phycobiliprotein photogenes in the unicellular rhodophyte, Cyanidium caldarium. J. Biol. Chem. 1989;264:20596–601. [PubMed] [Google Scholar]
- 44.Rhie G-e., Beale S.I. Regulation of heme oxygenase activity in Cyanidium caldarium by light, glucose, and phycobilin precursors. J. Biol. Chem. 1994;269:9620–6. [PubMed] [Google Scholar]
- 45.Kanesaki Y., Kobayashi Y., Hanaoka M., Tanaka K. Mg-protoporphyrin IX signaling in Cyanidioschyzon merolae: multiple pathways may involve the retrograde signaling in plant cells. Plant Signal. Behav. 2009;4:1190–2. doi: 10.4161/psb.4.12.10061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kobayashi Y., Imamura S., Hanaoka M., Tanaka K. A tetrapyrrole-regulated ubiquitin ligase controls algal nuclear DNA replication. Nat. Cell Biol. 2011;13:483–7. doi: 10.1038/ncb2203. [DOI] [PubMed] [Google Scholar]
- 47.Carter M.L., Smith A.C., Kobayashi H., Purton S., Herrin D.L. Structure, circadian regulation and bioinformatic analysis of the unique sigma factor gene in Chlamydomonas reinhardtii. Photosynth. Res. 2004;82:339–49. doi: 10.1007/s11120-004-4213-6. [DOI] [PubMed] [Google Scholar]
- 48.Bohne A.V., Irihimovitch V., Weihe A., Stern D.B. Chlamydomonas reinhardtii> encodes a single sigma70-like factor which likely functions in chloroplast transcription. Curr. Genet. 2006;49:333–40. doi: 10.1007/s00294-006-0060-7. [DOI] [PubMed] [Google Scholar]
- 49.Rochaix J.D. Chlamydomonas, a model system for studying the assembly and dynamics of photosynthetic complexes. FEBS Lett. 2002;529:34–8. doi: 10.1016/s0014-5793(02)03181-2. [DOI] [PubMed] [Google Scholar]
- 50.Choquet Y., Wollman F.A. Translational regulations as specific traits of chloroplast gene expression. FEBS Lett. 2002;529:39–42. doi: 10.1016/s0014-5793(02)03260-x. [DOI] [PubMed] [Google Scholar]
- 51.Allen J.F., Pfannschmidt T. Balancing the two photosystems: photosynthetic electron transfer governs transcription of reaction centre genes in chloroplasts. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2000;355:1351–9. doi: 10.1098/rstb.2000.0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matsuo M., Obokata J. Dual roles of photosynthetic electron transport in photosystem I biogenesis: light induction of mRNAs and chromatic regulation at post-mRNA level. Plant Cell Physiol. 2002;43:1189–97. doi: 10.1093/pcp/pcf146. [DOI] [PubMed] [Google Scholar]
- 53.Shao N., Vallon O., Dent R., Niyogi K.K., Beck C.F. Defects in the cytochrome b6/f complex prevent light-induced expression of nuclear genes involved in chlorophyll biosynthesis. Plant Physiol. 2006;141:1128–37. doi: 10.1104/pp.106.081059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hedtke B., Wagner I., Börner T., Hess W.R. Inter-organellar crosstalk in higher plants: impaired chloroplast development affects mitochondrial gene and transcript levels. Plant J. 1999;19:635–43. doi: 10.1046/j.1365-313x.1999.00554.x. [DOI] [PubMed] [Google Scholar]
- 55.Minoda A., Weber A.P., Tanaka K., Miyagishima S.Y. Nucleus-independent control of the Rubisco operon by the plastid-encoded transcription factor Ycf30 in the red alga Cyanidioschyzon merolae. Plant Physiol. 2010;154:1532–40. doi: 10.1104/pp.110.163188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang J., Schuster G., Stern D.B. CSP41, a sequence-specific chloroplast mRNA binding protein, is an endoribonuclease. Plant Cell. 1996;8:1409–20. doi: 10.1105/tpc.8.8.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bollenbach T.J., Stern D.B. Secondary structures common to chloroplast mRNA 3′-untranslated regions direct cleavage by CSP41, an endoribonuclease belonging to the short chain dehydrogenase/reductase superfamily. J. Biol. Chem. 2003;278:25832–8. doi: 10.1074/jbc.M303559200. [DOI] [PubMed] [Google Scholar]
- 58.Okuda K., Myouga F., Motohashi R., Shinozaki K., Shikanai T. Conserved domain structure of pentatricopeptide repeat proteins involved in chloroplast RNA editing. Proc. Natl Acad. Sci. USA. 2007;104:8178–83. doi: 10.1073/pnas.0700865104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matsuo T., Onai K., Okamoto K., Minagawa J., Ishiura M. Real-time monitoring of chloroplast gene expression by a luciferase reporter: evidence for nuclear regulation of chloroplast circadian period. Mol. Cell. Biol. 2006;26:863–70. doi: 10.1128/MCB.26.3.863-870.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Matsuoka K., Demura T., Galis I., et al. A comprehensive gene expression analysis toward the understanding of growth and differentiation of tobacco BY-2 cells. Plant Cell Physiol. 2004;45:1280–9. doi: 10.1093/pcp/pch155. [DOI] [PubMed] [Google Scholar]
- 61.Kuroiwa T., Fujie M., Kuroiwa H. Studies on the behavior of mitochondrial DNA. Synthesis of mitochondrial DNA occurs actively in a specific region just above the quiescent center in the root meristem of Pelargonium zonale. J. Cell Sci. 1992;101:483–93. [Google Scholar]
- 62.Maple J., Winge P., Tveitaskog A.E., Gargano D., Bones A.M., Møller S.G. Genome-wide gene expression profiles in response to plastid division perturbations. Planta. 2011;234:1055–63. doi: 10.1007/s00425-011-1459-z. [DOI] [PubMed] [Google Scholar]
- 63.Kuroiwa T. Mechanisms of organelle division and inheritance and their implications regarding the origin of eukaryotic cells. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2010;86:455–71. doi: 10.2183/pjab.86.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arimura S., Yamamoto J., Aida G.P., Nakazono M., Tsutsumi N. Frequent fusion and fission of plant mitochondria with unequal nucleoid distribution. Proc. Natl Acad. Sci. USA. 2004;101:7805–8. doi: 10.1073/pnas.0401077101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.