Abstract
Oncogenic BRAF mutations are found in several tumor types, including melanomas and colorectal cancers. Tumors with BRAF mutations have increased mitogen-activated protein kinase pathway activity and heightened sensitivity to BRAF and MEK (mitogen-activated or extracellular signal–regulated protein kinase kinase) inhibitors. To identify potential mechanisms of acquired drug resistance, we generated clones resistant to the allosteric MEK inhibitor AZD6244 from two BRAF V600E mutant colorectal cancer cell lines that are highly sensitive to MEK or BRAF inhibition. These AZD6244-resistant (AR) clones, which exhibited cross-resistance to BRAF inhibitors, acquired resistance through amplification of the BRAF gene. A small percentage of treatment-naïve parental cells showed preexisting BRAF amplification. We observed similar amplification in a subset of cells in a BRAF-mutant colorectal cancer. In cell lines, BRAF amplification increased the abundance of phosphorylated MEK and impaired the ability of AZD6244 to inhibit ERK (extracellular signal–regulated kinase) phosphorylation. The ability of AZD6244 to inhibit ERK phosphorylation in AR cells was restored by treatment with a BRAF inhibitor at low concentrations that reduced the abundance of phosphorylated MEK to amounts observed in parental cells. Combined MEK and BRAF inhibition fully overcame resistance to MEK or BRAF inhibitors alone and was also more effective in parental cells compared to treatment with either inhibitor alone. These findings implicate BRAF amplification as a mechanism of resistance to both MEK and BRAF inhibitors and suggest combined MEK and BRAF inhibition as a clinical strategy to overcome, or possibly prevent, this mechanism of resistance.
INTRODUCTION
Mutations in the BRAF proto-oncogene are found in many tumor types, including 40 to 60% of melanomas, 40% of thyroid cancers, and 10 to 20% of colorectal cancers. Most of these mutations encode a substitution of valine at amino acid 600 (V600) in BRAF (1). The presence of the BRAF V600 mutation predicts for sensitivity to inhibitors of the kinases MEK (mitogen-activated or extracellular signal–regulated protein kinase kinase) and BRAF in various preclinical models (2, 3). Consequently, these agents are being actively investigated in clinical trials. Although early trials with these drugs in unselected patient populations produced few responses (4, 5), recent clinical trials have focused on administering such inhibitors specifically to patients with BRAF-mutant tumors. This approach has yielded favorable response rates with the selective BRAF inhibitor PLX4032 and the allosteric MEK inhibitor GSK1120212 (6, 7). Other BRAF and MEK inhibitors are currently being developed for this patient population, and promising results are emerging.
Experience with similarly effective targeted therapies indicates that, despite marked initial responses, drug resistance frequently emerges, thereby limiting the clinical benefit of these drugs. Because BRAF and MEK inhibitors are still in early stages of clinical investigation, the small number of patients exposed to these drugs and the limited clinical samples available from these patients make it difficult to establish the mechanisms of resistance that may arise during treatment with these agents. However, pre-clinical modeling of acquired drug resistance has been useful for predicting the resistance mechanisms that emerge in patients receiving targeted cancer therapies, and these findings have led to strategies to overcome resistance that are now being used in the clinic (8–12).
In the case of BRAF-mutant tumors, preclinical models have identified two potential mechanisms of resistance to BRAF and MEK inhibitors. Increased CRAF activity was identified in drug-resistant clones derived from the highly sensitive BRAF V600E M14 melanoma cell line treated with the BRAF inhibitor AZ628 (13). Similarly, point mutations in MEK1 that conferred resistance to the MEK inhibitor AZD6244 were identified in the BRAF V600E A375 melanoma cell line. One of these point mutations was found in a drug-resistant focus of disease obtained from a patient with melanoma who had initially achieved stable disease with AZD6244 treatment (12).
Here, we used two highly sensitive BRAF-mutant colorectal cancer cell lines to model acquired resistance to a MEK inhibitor. We used methodologies that previously identified clinically validated mechanisms of resistance to targeted therapies (8–11). In both cell line models studied herein, BRAF gene amplification emerged as a robust mechanism of resistance to AZD6244 and also conferred cross-resistance to BRAF inhibitors. We observed that the signaling changes imparted by BRAF amplification altered the ability of AZD6244 to inhibit MEK-induced phosphorylation of extracellular signal–regulated kinase (ERK). However, we also determined that sensitivity to AZD6244 could be restored by co-treatment with subtherapeutic doses of the BRAF inhibitor AZ628. These studies implicate BRAF gene amplification as a potential mechanism of acquired resistance to MEK and BRAF inhibitors in tumors harboring the BRAF V600E mutation and offer potential therapeutic strategies to restore sensitivity.
RESULTS
AZD6244-resistant clones exhibit hyperactivation of the mitogen-activated protein kinase pathway
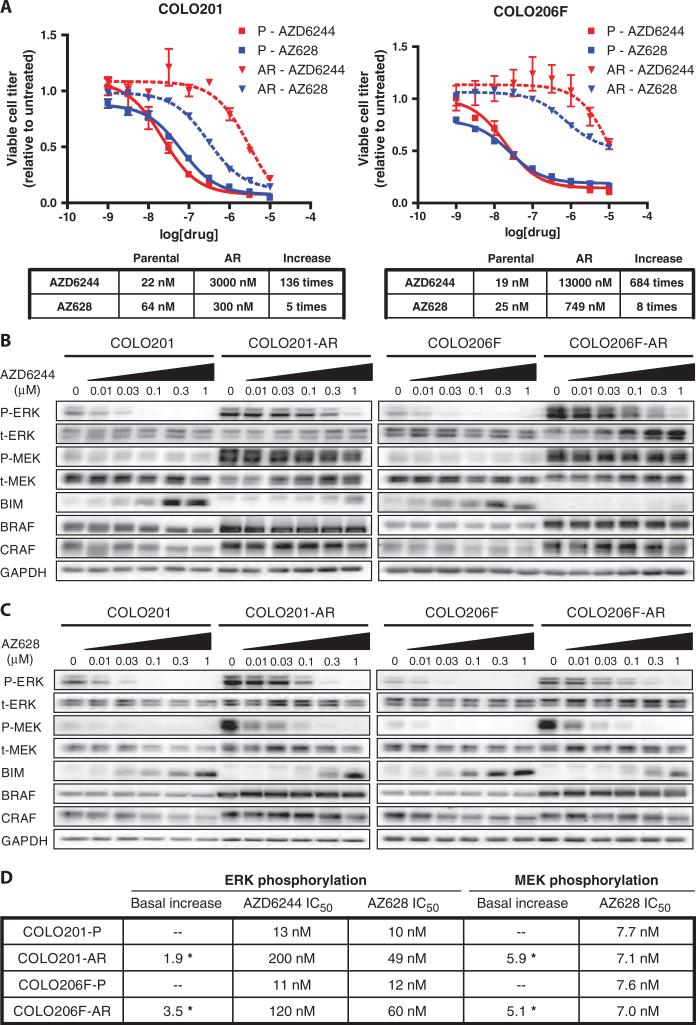
To identify potential mechanisms of acquired resistance to MEK inhibitors in BRAF-mutant tumors, we modeled resistance in vitro with two colorectal cancer cell lines, COLO201 and COLO206F. These cell lines harbor the BRAF V600E mutation and are sensitive to BRAF or MEK inhibitors, which decrease cell proliferation and induce apoptosis in these cell lines, leading to a reduction in viable cell titer (fig. S1, A and B). Cells were cultured in increasing concentrations of the allosteric MEK inhibitor AZD6244 until a pool of drug-resistant clones capable of proliferating in 1 μM AZD6244 was obtained for each cell line. The resulting AZD6244-resistant (AR) cells were termed COLO201-AR and COLO206F-AR. AR cells were more than 100 times less sensitive to AZD6244 than their parent lines and were also resistant to three additional MEK inhibitors (Fig. 1A and fig. S1C). AR cells also demonstrated cross-resistance to the selective BRAF inhibitors AZ628 and PLX4720 (Fig. 1A and fig. S1C).
Fig. 1.
AR clones are resistant to MEK and BRAF inhibition. (A) Parental (solid lines) COLO201 and COLO206F cells and AR (dashed lines) COLO201-AR and COLO206F-AR cells were treated in triplicate with the indicated concentrations of drug for 72 hours. Viable cell titer was determined, and the average values are shown relative to untreated controls for each cell line. Error bars represent the SD for each measurement. For each cell line, the IC50s for each inhibitor are shown in tabular form along with the increase in IC50 in AR cells relative to parental cells. (B and C) Western blots of RAF-MEK pathway components and effectors in parental and AR cells treated with the indicated concentrations of AZD6244 (B) or AZ628 (C) for 24 hours. (D) Tabular representation of chemiluminescent signal intensities from the blots in (B) and (C) showing IC50s for inhibition of ERK and MEK phosphorylation (full dose-response relationships are shown in fig. S2A). The statistically significant increases in basal phospho-ERK and phospho-MEK in AR cells relative to parental cells (average of at least three independent measurements) are also shown. *P < 0.01.
To evaluate the mechanism of resistance in each AR model, we assessed differences in signaling between parental and AR cells in response to MEK or BRAF inhibition. Changes in the mitogen-activated protein kinase (MAPK) signaling pathway were similar in both AR models, suggesting that a common resistance mechanism may have arisen in each. Compared to parental cells, basal ERK phosphorylation was increased in AR cells, and the ability of AZD6244 to inhibit ERK phosphorylation was attenuated (Fig. 1B). Indeed, ERK phosphorylation was detectable even in the presence of 1 μM AZD6244. Accordingly, in AR cells, AZD6244 failed to induce accumulation of the proapoptotic protein BIM (Bcl-2–interacting mediator of cell death), which is negatively regulated by ERK and has been implicated as the primary mediator of apoptosis in response to RAF or MEK inhibition (14). As expected, AZD6244 failed to induce marked apoptosis in AR cells (fig. S1B). The absolute amount of phosphorylated ERK (phospho-ERK) remaining after AZD6244 treatment correlated with BIM induction. For example, treatment of the COLO201-AR cells with 1 μM AZD6244 led to similar amounts of BIM as treatment with 30 nM AZD6244 of the parental cells (Fig. 1B). These data and the cell survival data suggest that the remaining absolute amount of phospho-ERK may be critical in determining the efficacy of a given dose of AZD6244 in inhibiting cell proliferation and inducing apoptosis.
We observed two potential factors that contributed to maintaining the absolute amount of phospho-ERK in the resistant cells despite treatment with AZD6244. First, the IC50 (median inhibitory concentration) of AZD6244 for inhibition of ERK phosphorylation was substantially increased in AR cells (Fig. 1, B and D, and fig. S2A). Second, because the basal abundance of phospho-ERK was higher in AR cells, a greater percent suppression of phospho-ERK was needed to decrease phospho-ERK to the same absolute amount as in parental cells. For example, a 50% reduction in ERK phosphorylation in parental COLO206F cells resulted in absolute amounts of phospho-ERK that were equal to an 87% reduction in the resistant cells (fig. S2A).
Basal phosphorylation of MEK was also markedly increased in AR cells, suggesting that signals contributing to the increased basal phosphorylation of ERK in AR cells were originating upstream, or at the level, of MEK (Fig. 1B). Therefore, we assessed the abundance of BRAF and CRAF, which phosphorylate MEK, and found that BRAF abundance was markedly increased. There was also a modest increase in CRAF abundance. Increased BRAF abundance appeared to be responsible for the hyperphosphorylation of MEK in AR cells, because treatment of AR cells with the BRAF inhibitor AZ628 completely inhibited MEK phosphorylation (Fig. 1C). The ability of AZ628 to inhibit phosphorylation of MEK by BRAF in AR cells was unaffected, as indicated by the unaltered IC50 of AZ628 for inhibition of MEK phosphorylation (Fig. 1D and fig. S2A). However, the ability of AZ628 to inhibit ERK phosphorylation was reduced (Fig. 1, C and D, and fig. S2, A and B), resulting in an increase in the IC50 for ERK phosphorylation (Fig. 1D). Because the basal amounts of phosphorylated MEK (phospho-MEK) in AR cells were more than five times higher than in parental cells, ~100 nM AZ628 is required to reduce phospho-MEK to amounts equivalent to those in the untreated parental cells (fig. S2A). As with AZD6244, the ability of AZ628 to inhibit cell viability mirrored its impact on the absolute amount of phospho-ERK.
Evaluation of the dose-response relationship between AZ628 and inhibition of the phosphorylation of MEK and ERK suggests that increased activation of MEK likely underlies the resistance to AZ628 observed in the AR cells. For example, in parental cells, 10 nM AZ628 reduced phospho-MEK abundance by ~50% and phospho-ERK abundance by ~50%. However, in AR cells, 10 nM AZ628 also reduced phospho-MEK by ~50%, but only reduced phospho-ERK by less than 15% (Fig. 1C and fig. S2B, pink dashed lines). In fact, to reduce phospho-ERK abundance by 50%, phospho-MEK abundance needed to be reduced by >85% in AR cells (fig. S2B, blue dashed lines). This observation suggests that in AR cells, increased BRAF abundance causes an excess of activated (phosphorylated) MEK, and substantially greater MEK inhibition (either directly by a MEK inhibitor, or indirectly by reducing the phosphorylation of MEK by a BRAF inhibitor) is required before leading to a decrease in ERK phosphorylation. This suggests that the amount of activated MEK is in excess of what is required for near-maximal ERK phosphorylation. Of note, this excess of activated MEK probably also contributes to the decreased effect of AZD6244 on ERK phosphorylation in the resistant cells (Fig. 1, B and D, and fig. S2A).
The BRAF gene is amplified in AR cells
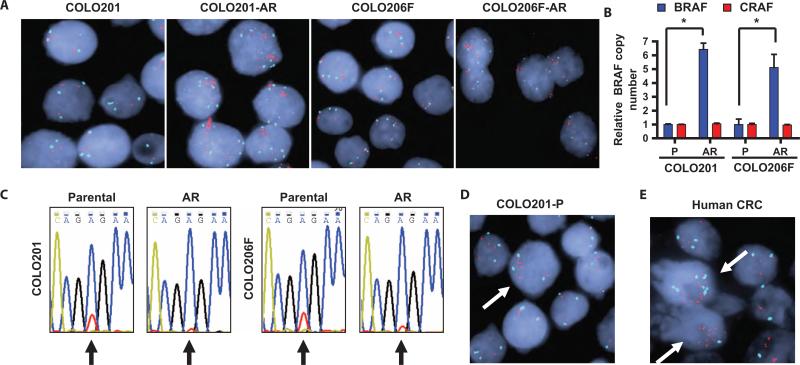
Because BRAF abundance was increased in the AR cells, we evaluated whether the BRAF gene was amplified. Fluorescence in situ hybridization (FISH) analysis showed a marked increase in BRAF gene copies in COLO201-AR and COLO206F-AR cells, relative to their respective parental cells (Fig. 2A). BRAF gene amplification was evident in 98 and 86% of COLO201-AR and COLO206F-AR cells, respectively, demonstrating that this molecular event is present in nearly all resistant cells. In contrast, there was no increase in CRAF gene copy number in AR cells, suggesting that the modest increases in CRAF abundance observed in AR cells reflected a distinct mechanism (fig. S3). To confirm and further quantify the degree of BRAF amplification, we performed quantitative polymerase chain reaction (PCR) from genomic DNA. This revealed that BRAF copy number was five to seven times greater in AR cells relative to parental cells (Fig. 2B). BRAF copy number was determined to be ~20 to 25 in COLO201-AR cells and ~10 to 15 in COLO206F cells. Quantitative PCR did not demonstrate an increase in CRAF DNA in AR cells, in agreement with the FISH data. Sequencing of parental and AR cells did not reveal any MEK1 mutations or new BRAF exon 15 mutations, but sequencing chromatograms showed that the peak height ratio of the mutant allele to the wild-type allele was greatly increased in the AR cells, suggesting selective amplification of the mutant BRAF allele (Fig. 2C).
Fig. 2.
AR cells have amplification of BRAF. (A) FISHimages showingincrease in BRAF copy number in AR cells relative to parental cells. BRAF probes are red, centromere 7 is aqua, and nuclear stain is blue. (B) Quantitative PCR for BRAF and CRAF from genomic DNA from parental (P) and AR cells. Values are shown relative to the parental cell line. *P < 0.001. (C) Sequencing chromatograms for BRAF from parental and AR cells, with the nucleotide color scheme: A (blue), C (gold), G (black), T (red). The T→A nucleotide substitution encoding the V600E mutation occurs at position 1799, which is indicated by an arrow. The wild-type allele (GTG) is represented by the red (T) peak at position 1799, and the mutant allele (GAG) is represented by the blue (A) peak. (D) FISH image showing an example of a cell (arrow) with preexisting low-level amplification of BRAF from the parental COLO201 cell line. (E) FISH image of a human colorectal cancer harboring a V600E BRAF mutation. Two cells with preexisting high-level BRAF amplification are indicated by arrows.
Notably, in both parental cell lines, occasional cells showed amplification of BRAF, suggesting that AR cells might arise by expansion of clones with preexisting amplification of BRAF (Fig. 2D). These cells represented 4% of COLO201 cells and 3.5% of COLO206F cells. We also evaluated 11 human colorectal cancer specimens known to harbor BRAF V600E mutations by FISH to determine whether similar populations of cells with preexisting BRAF amplification might exist in human tumors. In one tumor, we found that 28% of cells had substantial BRAF gene amplification (Fig. 2E and table S1). In this tumor, 10% of tumor cells had a BRAF copy number of 10 or greater, which is similar to the BRAF copy number found in AR cells, implying that these clones would likely be resistant to MEK or BRAF inhibitor therapy. This finding confirms that BRAF amplification occurs in human tumors harboring V600E mutations and supports the notion that BRAF amplification may exist before treatment in patients and has the potential to be a mechanism for either de novo resistance or acquired resistance to MEK or BRAF inhibitors.
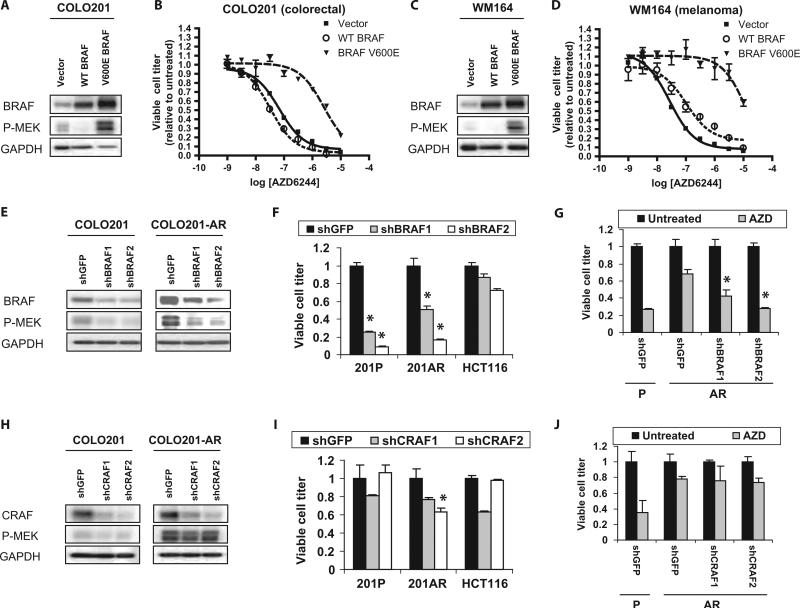
To determine whether increased BRAF abundance is sufficient to cause resistance to MEK inhibitors, we overexpressed either wild-type or mutant V600E BRAF in parental COLO201 cells. Similar to the AR cells, the COLO201 cells overexpressing V600E BRAF had increased amounts of phospho-MEK and demonstrated resistance to the effects of AZD6244 on viable cell titer (Fig. 3, A and B). Overexpression of V600E BRAF in a BRAF-mutated melanoma cell line, WM164, also led to increased phospho-MEK and resistance to MEK inhibitors (Fig. 3, C and D). This suggests that BRAF amplification could potentially lead to MEK inhibitor resistance in other BRAF-mutant cell lines and other tumor types. In contrast, overexpression of wild-type BRAF did not increase MEK phosphorylation or decrease sensitivity to MEK inhibition, consistent with the observation that AR cells specifically amplify the mutant BRAF allele (Fig. 2C). In fact, overexpression of wild-type BRAF appeared to modestly reduce phospho-MEK abundance, suggesting the possibility that the presence of large amounts of wild-type BRAF may interfere with the function of mutant BRAF.
Fig. 3.
Increased BRAF V600E is responsible for AZD6244 resistance in AR cells. (A and C) Western blots of COLO201 (A) and WM164 (C) cells infected with retroviruses encoding wild-type (WT) BRAF, mutant V600E BRAF, or vector control. (B and D) COLO201 (B) or WM164 (D) cells expressing WT BRAF, V600E BRAF, or vector control were treated with the indicated concentrations of AZD6244 for 72 hours and viable cell titer was determined. (E and H) Western blots of COLO201 and COLO201-AR cells infected with control shRNA (shGFP) or two different shRNAs directed against either BRAF (E) or CRAF (H). After infection, cells were selected in puromycin for 96 hours. Experiments were performed at least three times. (F and I) COLO201 (201P), COLO201-AR (201AR), and the KRAS mutant HCT116 cell line were infected with the above shRNAs. Cell viability was determined after 5 days of puromycin selection. Viral titers were used that yielded about the same results in the presence or absence of puromycin. *P < 0.001 relative to HCT116 cells infected with corresponding shRNA. (G and J) COLO201 (P) and COLO201-AR (AR) cells infected with the indicated shRNAs were selected in puromycin for 48 hours and then treated with or without 100 nM AZD6244 (AZD) for 72 hours before determination of cell viability. Viable cell titer is shown relative to the untreated control for each shRNA. *P < 0.001 relative to shGFP-infected COLO201-AR cells treated with AZD6244.
To test whether the marked amplification of BRAF observed in AR cells was responsible for the resistance to MEK inhibition we observed in these cells, we examined the consequences of BRAF-targeted short hairpin RNAs (shRNAs) in COLO201 and COLO201-AR cells. Both BRAF shRNAs tested led to a significant decrease in MEK phosphorylation (P < 0.001) in both COLO201 and COLO201-AR cells (Fig. 3E), suggesting that the increased MEK phosphorylation in AR cells is caused primarily by the amplified BRAF. As expected, parental COLO201 cells were highly BRAF-dependent, exhibiting a substantial reduction in viable cell number when treated with either BRAF shRNA (Fig. 3F). Similarly, BRAF knockdown reduced the titer of viable COLO201-AR cells, confirming that COLO201-AR cells indeed remain dependent on BRAF signaling. HCT116, a KRAS mutant colorectal cancer cell line that exhibits only modest sensitivity to BRAF and MEK inhibition, was used as a control for off-target toxicity of the shRNAs. As anticipated, only modest reductions in viable cell number were observed when this cell line was treated with the BRAF shRNAs.
BRAF knockdown restored the sensitivity of COLO201-AR cells to AZD6244 so that their sensitivity was comparable to that of parental COLO201 cells [infected with control short hairpin green fluorescent protein (shGFP) only] (Fig. 3G). In contrast, CRAF knockdown did not reduce the amount of phospho-MEK (Fig. 3H) and did not markedly reduce the viable cell number of parental or AR cells (Fig. 3I). One CRAF shRNA (shCRAF2) caused a reduction in viable cell titer in AR cells that was significant when compared to the effect of shCRAF2 in HCT116 cells. However, the magnitude of this reduction was small when compared to the effect of BRAF shRNA (84% reduction with shBRAF2 versus 37% reduction with shCRAF2; P < 0.001). Likewise, CRAF knockdown did not significantly increase the sensitivity of AR cells to AZD6244, suggesting that the slight increase in CRAF abundance seen in AR cells does not substantially contribute to MEK inhibitor resistance (Fig. 3J) Knockdown of BRAF, but not CRAF, also restored the ability of AZD6244 to decrease ERK phosphorylation and to induce BIM in AR cells (fig. S4). These results suggest that decreasing BRAF abundance and MEK activation so that they are comparable to those in the parental cells overcomes the resistance of AR cells to AZD6244.
Co-inhibition of BRAF and MEK restores sensitivity to AR cells
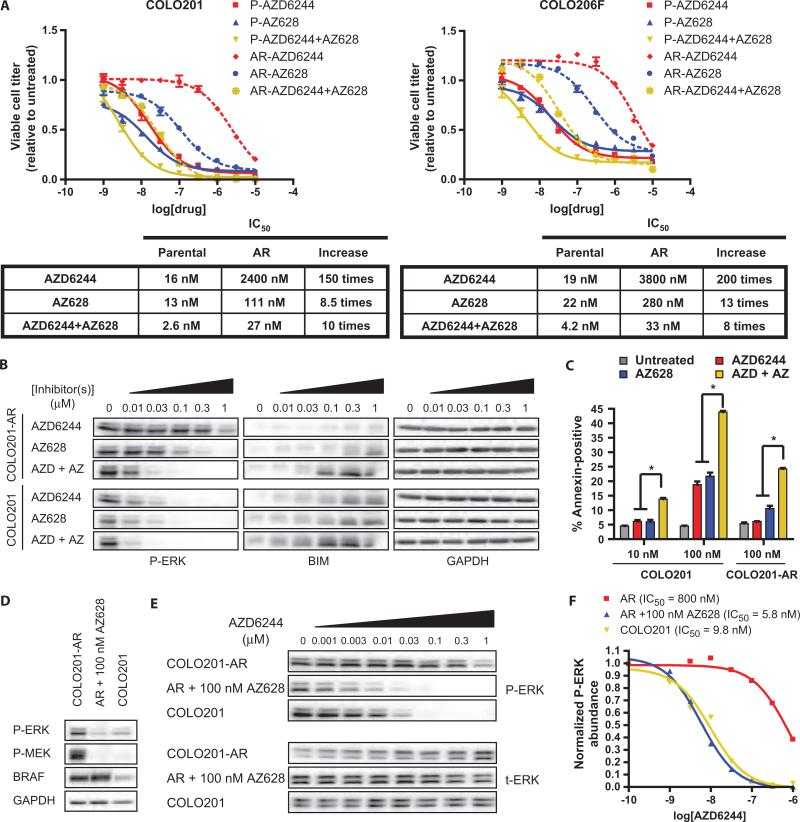
Because amplification of mutant BRAF in AR cells caused hyperactivation of MEK and resistance to AZD6244, we hypothesized that inhibiting excess BRAF activity might restore sensitivity to AZD6244. To test this hypothesis, we treated parental and AR cells with increasing concentrations of AZD6244 or the BRAF inhibitor AZ628, alone or in combination. Although AR cells were resistant to treatment with either compound alone, they were highly sensitive to the combination (Fig. 4A). In fact, the IC50s for the combination treatment in AR cells were similar to the IC50s of either inhibitor alone in parental cells. Moreover, parental COLO201 cells engineered to overexpress V600E BRAF were resistant to AZD6244 and AZ628, but were sensitive to the combination (fig. S5). The combination of AZD6244 and AZ628 also inhibited the parental cell lines more potently than did either treatment alone (Fig. 4A), suggesting that combinatorial targeting of the MAPK pathway may be an advantageous strategy in BRAF-mutant tumors, even in the absence of BRAF gene amplification.
Fig. 4.
Resistance to MEK and BRAF inhibition in AR cells can be overcome by combination treatment with AZD6244 and AZ628. (A) Parental (P, solid lines) and AR cells (dashed lines) were treated with the indicated concentrations of inhibitors for 72 hours and viable cell titer was determined. AZD6244 + AZ628 treatments contain the indicated concentrations of each inhibitor in combination. The IC50s for each inhibitor alone and in combination are shown in tabular form along with the increase in IC50 in AR cells relative to parental cells. (B) Western blot of COLO201 or COLO201-AR cells treated with the indicated concentrations of AZD6244 or AZ628 alone, or in combination (AZD + AZ) for 24 hours. (C) COLO201 or COLO201-AR cells were treated for 72 hours with the indicated concentrations of inhibitors. Percent of apoptotic cells was determined by annexin V staining. *P < 0.001 relative to treatment with either inhibitor alone. (D) Western blot of COLO201 cells and COLO201-AR cells treated with or without 100 nM AZ628 for 24 hours. Experiments were performed at least three times. (E) Western blot of COLO201 and COLO201-AR cells in the presence or absence of 100 nM AZ628 and treated with the indicated concentrations of AZD6244 for 24 hours. (F) Dose-response curves were generated by plotting normalized phospho-ERK (P-ERK) abundance versus AZD6244 concentration determined from chemiluminescence signal intensity measurements of the blots shown in (E). Phospho-ERK abundance was normalized to cells in the absence of AZD6244. Calculated IC50 values are also shown.
Combined MEK and BRAF inhibition also more potently decreased ERK phosphorylation in parental and AR cells (Fig. 4B), and again, a strong correlation between BIM induction and the absolute amount of phospho-ERK was observed. Consistent with these findings, we observed that the combination of AZD6244 and AZ628 enhanced the apoptotic response in parental and AR cells (Fig. 4C). At a concentration of 100 nM, either AZD6244 or AZ628 alone was sufficient to cause marked apoptosis in COLO201 cells. In contrast, at this same concentration, neither AZD6244 nor AZ628 alone caused a substantial increase in apoptosis in COLO201-AR cells. However, when these agents were combined at 100 nM each, we observed an increase in apoptosis in the AR cells that was equivalent to that induced by either agent alone in parental COLO201 cells. Similarly, in parental COLO201 cells, the combination of AZD6244 and AZ628 induced considerably more apoptosis than equal concentrations of either agent alone. In fact, the combination of 10 nM AZD6244 and 10 nM AZ628 induced nearly as much apoptosis as 100 nM of either agent alone. Collectively, these findings suggest that the combination of BRAF and MEK inhibition can not only overcome the resistance caused by BRAF amplification but also potentially enhance antitumor efficacy against BRAF-mutant tumors in general and allow for lower effective doses of each drug, regardless of BRAF amplification status.
Modulation of BRAF activity can alter the ability of AZD6244 to inhibit ERK phosphorylation
The IC50 of AZD6244 for inhibition of ERK phosphorylation was markedly increased in AR cells relative to parental cells (Fig. 1D and fig. S2A). Because combined inhibition of BRAF and MEK overcame the resistance of AR cells to either MEK or BRAF inhibitor alone, we tested whether inhibition of BRAF could restore the dose-response relationship between AZD6244 and inhibition of ERK phosphorylation in AR cells. We hypothesized that if the increased IC50 of AZD6244 for inhibition of ERK phosphorylation is due to increased MEK activation, it might be reversed in the presence of a concentration of BRAF inhibitor sufficient to decrease phospho-MEK to levels equivalent to those in parental cells. We used 100 nM AZ628, which reduced phospho-MEK abundance in COLO201-AR cells, so that it was comparable to the amount of basal phospho-MEK in COLO201 cells (Fig. 1C and fig. S2A). In COLO201-AR cells treated with 100 nM AZ628, phosphorylation of MEK and ERK was similar to that in untreated parental COLO201 cells (Fig. 4D). At this concentration, AZ628 completely restored the ability of AZD6244 to inhibit ERK phosphorylation in AR cells (Fig. 4E). Indeed, 100 nM AZ628 decreased the IC50 of AZD6244 for ERK phosphorylation in COLO201-AR cells by >100 times, so that the IC50s of AZD6244 for ERK phosphorylation in AZ628-treated COLO201-AR cells and parental COLO201 cells were virtually identical (Fig. 4F). These results show that increasing or decreasing BRAF activity (and consequently, phosphorylation and activation of MEK) can lead to a decrease or increase, respectively, in the ability of AZD6244 to inhibit MEK-mediated phosphorylation of ERK. By affecting the ability of AZD6244 to inhibit ERK phosphorylation, inhibition of BRAF activity and MEK activation can therefore critically enhance the antitumor efficacy of AZD6244.
We also evaluated the effect of AZ628 co-treatment on the capacity of AZD6244 to inhibit ERK phosphorylation in parental cells (fig. S6). Although no substantial change in the IC50 of AZD6244 was noted in the presence of AZ628 (fig. S6, B and C), the reduction in the absolute amount of ERK phosphorylation was much greater at a given concentration of AZD6244 (fig. S6, B and D). Therefore, enhanced inhibition of ERK phosphorylation likely underlies the increased potency with which the combination treatment inhibits cell viability in parental cells.
DISCUSSION
The administration of targeted therapies to patients whose cancers harbor specific genetic abnormalities has shown considerable promise (15). However, these therapies have frequently been limited by the eventual emergence of drug resistance. Because targeted therapies directed against BRAF-mutant tumors, such as BRAF and MEK inhibitors, have only recently entered clinical testing, there is minimal experience and limited clinical specimens from which to ascertain the major mechanisms of resistance that will arise in patients treated with these agents. Therefore, preclinical models can provide valuable tools to predict likely mechanisms of resistance to these agents and to guide clinical investigation so that the mechanisms of drug resistance that emerge in the clinic can be more efficiently identified, understood, and eventually overcome. Through in vitro modeling of drug resistance, we have identified BRAF gene amplification as a potential mechanism of acquired resistance to MEK inhibitors in tumors harboring the BRAF V600E mutation. Furthermore, BRAF amplification also caused cross-resistance to selective BRAF inhibitors, raising the possibility that patients receiving BRAF inhibitors might also develop this potential resistance mechanism.
Intriguingly, in parental cell populations, occasional cells with preexisting low degrees of BRAF amplification were noted. It is possible that these cells might have a selective advantage in the presence of MEK inhibitor and may serve as the founder cells for the eventual drug-resistant clones with high degrees of BRAF amplification that emerge after extended exposure to drug. Patients with EGFR (epidermal growth factor receptor)–mutated lung cancers who exhibit rare cells (often <1%) with preexisting MET amplification in their pretreatment biopsies are more likely to develop MET gene amplification as the eventual resistance mechanism to EGFR-directed therapy with erlotinib than patients without any detectable cells with preexisting MET amplification (16). Similarly, evaluation of pre-treatment biopsies of patientswith BRAF-mutant tumors might reveal those patients who are likely to develop BRAF amplification in response to MEK inhibitor therapy. Alternatively, the presence of more widespread gains in BRAF gene copy number at the time of diagnosis might identify a population of patients who are less likely to have a meaningful response to single-agent MEK or BRAF inhibitor and who may benefit from an alternative treatment regimen, such as a MEK and BRAF inhibitor combination.
The prevalence of BRAF copy number gains in tumors harboring the BRAF V600E mutation has not been extensively studied, but studies have identified BRAF copy number gains in human tumors, including melanoma and colorectal cancer (17, 18). We identified BRAF amplification as the primary resistance mechanism in both the COLO201 and the COLO206F models, suggesting that it may prove to be a common mode of resistance among BRAF-mutant tumors treated with this drug class. However, although COLO201 and COLO206F are independently established cell lines, they did originate from the same patient (19). Thus, we examined BRAF-mutated human colorectal cancer and identified BRAF amplification in 1 of 11 BRAF-mutated colorectal cancers evaluated by FISH. Twenty-eight percent of cells displayed BRAF amplification, and 10% of cells displayed amplification of 10 or more copies, similar to that seen in the AR cell lines. It is therefore likely that these tumor cells would be resistant to MEK or BRAF inhibitor therapy. Although we did not detect clones with preexisting BRAF amplification in the other 10 tumors examined, our methods would have failed to detect amplification events present in less than ~2% of cells. Thus, it is possible that this cohort may have included additional cancers with a small proportion of cells harboring BRAF amplification. Our results suggest that evaluation of BRAF copy number in patients participating in clinical trials of BRAF or MEK inhibitors might provide useful information to help predict patient response to therapy, although sensitive techniques capable of detecting small numbers of cells with preexisting amplification might be required.
Mechanisms of acquired drug resistance to targeted therapies commonly involve either mutations or amplifications of the drug target itself or changes unrelated to the drug target that activate parallel or downstream signaling pathways to circumvent the activity of the drug. For example, the T790M mutation in EGFR causes resistance to erlotinib, and point mutations in or amplifications of the BCR-ABL gene can produce resistance to imatinib (20–22). Similarly, a mutation in the MEK1 gene was recently identified in an AR disease focus of a patient with BRAF V600E mutant melanoma (12). Activation of parallel signaling pathways such as METand the insulin-like growth factor signaling axis can cause resistance to EGFR-directed therapies (10, 16, 23). Likewise, increased CRAF activity can cause resistance to BRAF inhibitors in BRAF-mutant cancer cells (13). However, the mechanism of resistance to MEK inhibition identified in this study is unusual in that it involves amplification of an upstream signaling component (BRAF) that leads to hyperactivation of the drug target itself (MEK) and thereby reduces the ability of AZD6244 to inhibit MEK-mediated ERK phosphorylation. It is interesting that BRAF amplification is ultimately able to achieve the same effect as a MEK point mutation (12), as each decreases the ability of AZD6244 to inhibit its target. Indeed, both BRAF amplification and the P124 MEK1 mutation identified by Emery et al. led to an ~10- to 100-fold increase in the amount of MEK inhibitor required for inhibition of ERK phosphorylation (12).
BRAF amplification appears to cause resistance to MEK and BRAF inhibitors through an excess of activated MEK, which has two important consequences: (i) an increase in the IC50 for inhibition of ERK phosphorylation and (ii) an increase in the basal amount of phospho-ERK. Furthermore, the studies with the BRAF inhibitor suggest that MEK is activated in the resistant cells in far excess of that needed for maximal ERK phosphorylation. For example, in AR cells, 10 nM AZ628 reduced the amount of phospho-MEK by ~50% but reduced phospho-ERK by less than 15% (fig. S2B). The effectiveness of the MEK inhibitors correlated with the reduction of absolute phospho-ERK, indicating that BRAF amplification and MEK hyperactivation conspire to maintain increased ERK activation in the presence of AZD6244 and produce a shift in the IC50 for cell viability that is substantially larger than the shift in the IC50 for inhibition of ERK phosphorylation alone.
Additional mechanisms may contribute to the shift of the IC50 of AZD6244 for inhibition of ERK phosphorylation in the resistant cells (Fig. 1D and fig. S2A). For example, it is possible that AZD6244 has a lower affinity for activated MEK than it does for inactive MEK. AZD6244 is an allosteric inhibitor that binds to a pocket adjacent to the activation loop of MEK, and it functions by binding and stabilizing the closed, inactive conformation of the enzyme (24). In the presence of BRAF amplification and the resulting MEK hyperactivation, if there is a large excess of activated MEK and relatively little MEK in the “preferred” inactive conformation, the ability of AZD6244 to bind to MEK may be decreased. Overcoming this decreased binding affinity for its target would require a higher concentration of drug to effectively bind and inhibit MEK, potentially accounting for the large increase in the IC50 of AZD6244 for the inhibition of MEK-mediated ERK phosphorylation in AR cells. In this scenario, when AR cells are co-treated with AZ628 and the fraction of inactive MEK increases, the proportion of MEK with high affinity for AZD6244 would be restored, and the dose-response relationship with ERK phosphorylation would shift to the left toward that of the parental cells, as was observed (Fig. 4, E and F).
It is intriguing to speculate that BRAF amplification is not the only change that could lead to hyperactivation of MEK and decreased potency of MEK and BRAF inhibitors. It is possible that excessive upstream input from CRAF, RAS proteins, or even receptor tyrosine kinases could similarly decrease the potency of MEK and BRAF inhibitors in BRAF wild-type tumors if sufficient MEK hyperactivation is achieved. Indeed, although an increase in basal phospho-MEK, such as seen with the BRAF V600E mutation, can be a marker for cells susceptible to MEK inhibition, it is possible that excessive phospho-MEK could paradoxically lead to decreased sensitivity.
Finally, our results provide a rationale for the investigational use of BRAF and MEK inhibitor combinations in patients with BRAF-mutant tumors. First, combination treatment with MEK and BRAF inhibitors may be useful in preventing emergence of resistance or in overcoming resistance to therapies targeting RAF or MEK. All three reported mechanisms of acquired resistance to MEK or BRAF inhibitors retain sensitivity to the combination of MEK and BRAF inhibition. MEK1 mutants retain sensitivity to the combination despite causing resistance to each drug individually (12). BRAF-mutant cancer cells associated with increased CRAF activity retain some sensitivity to MEK inhibition, although at reduced potency (13). Similarly, we show here that tumors with acquired amplification of BRAF V600E are as sensitive to combined MEK and BRAF inhibition as their treatment-naïve parental cells are to each drug individually. Therefore, combinatorial targeting of the RAF-MEK pathway may help to overcome or prevent these resistance mechanisms. Second, even in treatment-naïve parental cells, combined MEK and BRAF inhibition was about five times more potent than either agent alone (Fig. 4A). We found that this combination did not substantially change the IC50 for ERK phosphorylation in parental cells as it did for AR cells. However, it did markedly increase the degree of absolute inhibition of ERK phosphorylation achieved at a given dose of the combination compared to the same concentration of either drug alone. Of course, the combination of two drugs does have the potential to increase toxicity, but because the combination required substantially lower doses of each drug, it is possible that this approach could actually decrease toxicity. Lower concentrations of each drug needed for combination therapy could potentially reduce off-target toxicities of these agents, although there may be little difference in the on-target toxicity due to RAF-MEK pathway inhibition. Moreover, the lower concentrations of each drug required for the combination may be easier to achieve in patients. Therefore, we believe that combination therapy with MEK and BRAF inhibitors for tumors harboring BRAF V600E mutations presents an attractive strategy for clinical investigation.
MATERIALS AND METHODS
Cell lines, reagents, and patient samples
COLO201 and HCT116 cells were obtained from the American Type Culture Collection. COLO206F cells were obtained from DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH). WM164 cells were obtained from the Massachusetts General Hospital Center for Molecular Therapeutics. COLO201 and COLO206F were maintained and assayed in RPMI 1640 (Gibco) with 5% fetal bovine serum (FBS). HCT116 and WM164 cells were maintained and assayed in Dulbecco's modified Eagle's medium (DMEM)/F12 (Gibco) with 5% FBS. AZD6244 was purchased from Otava Chemicals. AZ628 was provided by AstraZeneca. PD0325901, CI-1040, U0126, and PLX4720 were purchased from Selleck Chemicals. All compounds were dissolved in dimethyl sulfoxide. Human colorectal cancer specimens were obtained from the Massachusetts General Hospital under institutional review board–approved studies. All patients provided written, informed consent. BRAF mutation status was determined by the Massachusetts General Hospital Clinical Laboratory and Department of Pathology.
Generation of AR clones
COLO201 and COLO206F cells were seeded at ~70% confluence in 10-cm plates in RPMI 1640 with 5% FBS. AZD6244 was added at a starting concentration of 10 nM, and cells were maintained in fresh drug-containing medium changed every ~72 hours. Cells were passaged once they reached ~70% confluence. After every two passages at a given concentration of drug, the concentration of AZD6244 was increased in half-log intervals until a final concentration of 1 μM was achieved. The resulting AR pooled clones were termed COLO201-AR and COLO206F-AR and were maintained in RPMI with 5% FBS containing 1 μM AZD6244.
Determination of viable cell titer
Cells were seeded at 2000 cells per well of a 96-well plate. After overnight incubation, the cells were treated in triplicate with serial dilutions of each drug for 72 hours. Viable cell titer relative to untreated cells was determined with CellTiter-Glo assay (Promega) according to the manufacturer's protocol and read on a Centro LB 960 microplate luminometer (Berthold Technologies). The CellTiter-Glo assay measures the titer of live, metabolically active cells in culture by quantification of the amount of adenosine 5′-triphosphate (ATP) present. This system has been widely used previously to assess the response of cancer cell lines to therapeutics, including BRAF-mutant cell lines treated with MEK and BRAF inhibitors (12, 25–27).
Annexin V apoptosis assays
Cells were seeded at ~30 to 40% confluence in 6-cm plates. After overnight incubation, the medium was aspirated and replaced with medium with or without various concentrations of indicated drugs. After 72 hours, the medium was collected. Cells were washed with phosphate-buffered saline (PBS) and trypsinized. PBS wash and trypsinized cells were added to the collected medium in a single tube. Cells were pelleted, washed once with PBS, and resuspended in Annexin binding buffer (BD Biosciences) at ~1 × 106 cells/ml. Cells were stained with propidium iodide (BD Biosciences) and Annexin V Cy5 (BioVision) according to the manufacturer's protocol and assayed on an LSRII flow cytometer (BD Biosciences).
Western blot analysis and quantification of chemiluminescent signal intensity
Western blotting was performed with standard methods. After 24 hours of treatment with indicated drugs, cells were washed with cold PBS and lysed in the following lysis buffer: 20 mM tris (pH 7.4), 150 mM NaCl, 1% NP-40, 10% glycerol, 1 mM EDTA, 1 mM EGTA, 5 mM sodium pyrophosphate, 50 mM NaF, 10 nM β-glycerophosphate, 1 mM sodium vanadate, 0.5 mM dithiothreitol, leupeptin (4 μg/ml), pepstatin (4 μg/ml), aprotinin (4 μg/ml), and 1 mM phenylmethylsulfonyl fluoride. Lysates were centrifuged at 16,000g for 5 min at 4°C. Protein concentrations were determined by BCA assay (Thermo Scientific). Proteins were resolved by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene difluoride membrane (Hybond-P, Amersham). Immunoblotting was performed per the antibody manufacturer's specifications. Antibodies to phospho-ERK1/2, total ERK1/2, phospho-MEK1/2, total MEK1/2, and BIM were purchased from Cell Signaling. Antibodies to BRAF and CRAF were purchased from Santa Cruz Biotechnology. Antibody to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was purchased from Chemicon. Chemiluminescence was detected with the Syngene G:Box camera (Synoptics), and chemiluminescent signal intensity was quantified with Syngene Genetools software (Synoptics). All measurements were performed in the linear range without saturation and were normalized to GAPDH loading control.
Isolation of genomic DNA and DNA sequencing
Genomic DNA was isolated from pelleted cells with a DNeasy kit (QIAGEN) according to the manufacturer's protocol. Exon 15 of BRAF and exons 3 and 6 of MEK1 were PCR-amplified from genomic DNA and sequenced bi-directionally by Sanger dideoxynucleotide sequencing with the following primers: BRAF, forward 5′-TCATAATGCTTGCTCTGATAGGA-3′, reverse 5′-CTTTCTAGTAACTCAGCAGC-3′; MEK1 exon 3, forward 5′-CTTTCATCCCTTCCTCCCTC-3′,reverse5′-CACCTCCCAGACCAAAGATTAG-3′; and MEK1 exon 6, forward 5′-CTTCTCTTCCCCAATCTACCTGTG-3′, reverse 5′-CCTACCCAGCACAAGACTCTG-3′.
Quantitative PCR
Quantitative PCRs were carried out in quadruplicate in 50-μl reactions containing 50 μg of genomic DNA with SYBR green master mix (Roche Diagnostics). Quantitative measurements were performed with a LightCycler 480 (Roche Diagnostics) under the following cycling conditions: 95°C for 10 min, followed by 35 cycles of 95°C for 15 s, 55°C for 60 s, and 72°C for 30 s. LINE-1 (long interspersed nuclear element 1) elements were used as a normalization control for DNA content. Copy number was calculated as previously described (17) with a standard curve of normal female genomic DNA (Promega). The following primers were used at a final concentration of 0.3 μM: LINE-1, forward 5′-AAAGCCGCTCAACTACATGG-3′, reverse 5′-TGCTTTGAATGCGTCCCAGAG-3′; BRAF, forward 5′-CAAGTCACCACAAAAACCTATCGT-3′, reverse 5′-AACTGACTCACCACTGTCCTCTGTT-3′; and CRAF, forward 5′-CAACTGATTGCACTGACTGCCAAC-3′, reverse 5′-CCAGCTTTCTACTCACCGCACAAC-3′.
Fluorescence in situ hybridization
Trypsinized cells were pelleted and fixed in 3:1 methanol/acetic acid, and FISH staining for BRAF and CRAF was performed as previously described (13). Cells were considered to have BRAF gene amplification if they had six or more copies of BRAF in addition to a BRAF–to–chromosome 7 ratio of 1.5 or greater. Quantification of gene copy number was based on individual cell counts of 200 cells for cell line experiments and 50 cells for human tumors.
Lentiviral shRNA experiments
shRNA constructs in the pLKO.1 lentiviral vector containing the following targeting sequences were used: shGFP, 5′-GCAAGCTGACCCTGAAGTTCAT-3′; shBRAF1, 5′-GCAGATGAAGATCATCGAAAT-3′; shBRAF2, 5′-CAGCAGTTACAAGCCTTCAAA-3′; shCRAF1, 5′-CATGAGTATTTAGAGGAAGTA-3′; and shCRAF2, 5′-GCTTCCTTATTCTCACATCAA-3′. Lentiviral particles were generated and target cells were infected as described previously (28). The day before infection, COLO201 and COLO201-AR cells were seeded in 96-well plates in triplicate at 2000 cells per well for cell viability assays and in 6-well plates at 3 × 105 cells per well for Western blot analysis. HCT116 cells were seeded in 96-well plates in triplicate at 1500 cells per well. The morning after infection, cells were treated with puromycin (2 μg/ml) for 48 to 72 hours to eliminate uninfected cells. Medium without puromycin containing the indicated concentrations of drug was then added for 72 hours for cell viability assays or 24 hours for Western blot analysis.
BRAF expression constructs
Constructs encoding wild-type and V600E mutant BRAF in the pBABE-puro vector were a gift of C. Der and J. Shields, University of North Carolina, Chapel Hill (29). Constructs were transfected into Phoenix Ampho Cells (Orbigen) according to the manufacturer's protocol. Target cells were seeded in six-well plates at a density of 4 × 104 to 12 × 104 cells per well the day before infection. Retroviral supernatants were applied to target cells in the presence of polybrene (8 μg/ml), and plates were centrifuged at 32°C and 2250 rpm for 1.5 hours. Medium containing puromycin (2 μg/ml) was added to cells the following morning. After 4 days of puromycin selection, cells were assayed by Western blot or used for cell viability assays.
Determination of IC50 and statistical analysis
Statistical analyses were performed with a two-tailed Student's t test (Fig. 1D) or analysis of variance (ANOVA) with Bonferroni's post test (Figs. 2 to 4). Significance was established for P value <0.01. IC50 and GI50 (defined as the concentration required to inhibit viable cell titer to 50% of untreated control) determinations were made with GraphPad Prism software.
Supplementary Material
Acknowledgments
We thank C. Der and J. Shields for providing BRAF expression constucts. Funding: R.B.C. was supported by NIH training grant T32 CA071345. This study was supported by NIH grants K08 CA120060-01 (to J.A.E.), R01CA137008-01 (to J.A.E.), R01CA140594 (to J.A.E.), and R01CA135257-01 (to J.A.E.); National Cancer Institute Lung Specialized Program of Research Excellence (SPORE) grant P50CA090578 (to J.A.E.); Dana-Farber/Harvard Cancer Center Gastrointestinal Cancer SPORE grant P50 CA127003 (to J.A.E.); the American Association for Cancer Research (to J.A.E.); the V Foundation (to J.A.E.); American Cancer Society grant RSG-06-102-01-CCE (to J.A.E.); and the Ellison Foundation Scholar (to J.A.E.).
Footnotes
Author contributions: R.B.C. performed the experiments; K.B., D.D.-S., and A.J.I. performed and analyzed the FISH experiments; R.B.C., J.A.E., and J.S. designed the experiments, analyzed the data, and wrote the manuscript.
SUPPLEMENTARY MATERIALS
Citation: R. B. Corcoran, D. Dias-Santagata, K. Bergethon, A. J. Iafrate, J. Settleman, J. A. Engelman, BRAF gene amplification can promote acquired resistance to MEK inhibitors in cancer cells harboring the BRAF V600E mutation. Sci. Signal. 3, ra84 (2010).
REFERENCES AND NOTES
- 1.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JW, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 2.Solit DB, Garraway LA, Pratilas CA, Sawai A, Getz G, Basso A, Ye Q, Lobo JM, She Y, Osman I, Golub TR, Sebolt-Leopold J, Sellers WR, Rosen N. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006;439:358–362. doi: 10.1038/nature04304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McDermott U, Sharma SV, Dowell L, Greninger P, Montagut C, Lamb J, Archibald H, Raudales R, Tam A, Lee D, Rothenberg SM, Supko JG, Sordella R, Ulkus LE, Iafrate AJ, Maheswaran S, Njauw CN, Tsao H, Drew L, Hanke JH, Ma XJ, Erlander MG, Gray NS, Haber DA, Settleman J. Identification of genotype-correlated sensitivity to selective kinase inhibitors by using high-throughput tumor cell line profiling. Proc. Natl. Acad. Sci. U.S.A. 2007;104:19936–19941. doi: 10.1073/pnas.0707498104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rinehart J, Adjei AA, Lorusso PM, Waterhouse D, Hecht JR, Natale RB, Hamid O, Varterasian M, Asbury P, Kaldjian EP, Gulyas S, Mitchell DY, Herrera R, Sebolt-Leopold JS, Meyer MB. Multicenter phase II study of the oral MEK inhibitor, CI-1040, in patients with advanced non-small-cell lung, breast, colon, and pancreatic cancer. J. Clin. Oncol. 2004;22:4456–4462. doi: 10.1200/JCO.2004.01.185. [DOI] [PubMed] [Google Scholar]
- 5.Adjei AA, Cohen RB, Franklin W, Morris C, Wilson D, Molina JR, Hanson LJ, Gore L, Chow L, Leong S, Maloney L, Gordon G, Simmons H, Marlow A, Litwiler K, Brown S, Poch G, Kane K, Haney J, Eckhardt SG. Phase I pharmacokinetic and pharmacodynamic study of the oral, small-molecule mitogen-activated protein kinase kinase 1/2 inhibitor AZD6244 (ARRY-142886) in patients with advanced cancers. J. Clin. Oncol. 2008;26:2139–2146. doi: 10.1200/JCO.2007.14.4956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, O'Dwyer PJ, Lee RJ, Grippo JF, Nolop K, Chapman PB. Inhibition of mutated, activated BRAF in metastatic melanoma. N. Engl. J. Med. 2010;363:809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Infante JR, Fecher LA, Nallapareddy S, Gordon MS, Flaherty KT, Cox DS, DeMarini DJ, Morris SR, Burris HA, Messersmith WA. Safety and efficacy results from the first-in-human study of the oral MEK 1/2 inhibitor GSK1120212, 2010 ASCO Annual Meeting; American Society of Clinical Oncology, Alaxandria, VA. 2010; Abstract no. 2503. [Google Scholar]
- 8.Azam M, Latek RR, Daley GQ. Mechanisms of autoinhibition and STI-571/imatinib resistance revealed by mutagenesis of BCR-ABL. Cell. 2003;112:831–843. doi: 10.1016/s0092-8674(03)00190-9. [DOI] [PubMed] [Google Scholar]
- 9.Engelman JA, Mukohara T, Zejnullahu K, Lifshits E, Borrás AM, Gale CM, Naumov GN, Yeap BY, Jarrell E, Sun J, Tracy S, Zhao X, Heymach JV, Johnson BE, Cantley LC, Jänne PA. Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR-amplified lung cancer. J. Clin. Invest. 2006;116:2695–2706. doi: 10.1172/JCI28656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, Kosaka T, Holmes AJ, Rogers AM, Cappuzzo F, Mok T, Lee C, Johnson BE, Cantley LC, Jänne PA. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 11.Godin-Heymann N, Ulkus L, Brannigan BW, McDermott U, Lamb J, Maheswaran S, Settleman J, Haber DA. The T790M “gatekeeper” mutation in EGFR mediates resistance to low concentrations of an irreversible EGFR inhibitor. Mol. Cancer Ther. 2008;7:874–879. doi: 10.1158/1535-7163.MCT-07-2387. [DOI] [PubMed] [Google Scholar]
- 12.Emery CM, Vijayendran KG, Zipser MC, Sawyer AM, Niu L, Kim JJ, Hatton C, Chopra R, Oberholzer PA, Karpova MB, MacConaill LE, Zhang J, Gray NS, Sellers WR, Dummer R, Garraway LA. MEK1 mutations confer resistance to MEK and B-RAF inhibition. Proc. Natl. Acad. Sci. U.S.A. 2009;106:20411–20416. doi: 10.1073/pnas.0905833106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Montagut C, Sharma SV, Shioda T, McDermott U, Ulman M, Ulkus LE, Dias-Santagata D, Stubbs H, Lee DY, Singh A, Drew L, Haber DA, Settleman J. Elevated CRAF as a potential mechanism of acquired resistance to BRAF inhibition in melanoma. Cancer Res. 2008;68:4853–4861. doi: 10.1158/0008-5472.CAN-07-6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wickenden JA, Jin H, Johnson M, Gillings AS, Newson C, Austin M, Chell SD, Balmanno K, Pritchard CA, Cook SJ. Colorectal cancer cells with the BRAFV600E mutation are addicted to the ERK1/2 pathway for growth factor-independent survival and repression of BIM by BRAFV600E. Oncogene. 2008;27:7150–7161. doi: 10.1038/onc.2008.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McDermott U, Settleman J. Personalized cancer therapy with selective kinase inhibitors: An emerging paradigm in medical oncology. J. Clin. Oncol. 2009;27:5650–5659. doi: 10.1200/JCO.2009.22.9054. [DOI] [PubMed] [Google Scholar]
- 16.Turke AB, Zejnullahu K, Wu YL, Song Y, Dias-Santagata D, Lifshits E, Toschi L, Rogers A, Mok T, Sequist L, Lindeman NI, Murphy C, Akhavanfard S, Yeap BY, Xiao Y, Capelletti M, Iafrate AJ, Lee C, Christensen JG, Engelman JA, Jänne PA. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Modrek B, Ge L, Pandita A, Lin E, Mohan S, Yue P, Guerrero S, Lin WM, Pham T, Modrusan Z, Seshagiri S, Stern HM, Waring P, Garraway LA, Chant J, Stokoe D, Cavet G. Oncogenic activating mutations are associated with local copy gain. Mol. Cancer Res. 2009;7:1244–1252. doi: 10.1158/1541-7786.MCR-08-0532. [DOI] [PubMed] [Google Scholar]
- 18.Soh J, Okumura N, Lockwood WW, Yamamoto H, Shigematsu H, Zhang W, Chari R, Shames DS, Tang X, MacAulay C, Varella-Garcia M, Vooder T, Wistuba II, Lam S, Brekken R, Toyooka S, Minna JD, Lam WL, Gazdar AF. Oncogene mutations, copy number gains and mutant allele specific imbalance (MASI) frequently occur together in tumor cells. PLoS One. 2009;4:e7464. doi: 10.1371/journal.pone.0007464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Semple TU, Quinn LA, Woods LK, Moore GE. Tumor and lymphoid cell lines from a patient with carcinoma of the colon for a cytotoxicity model. Cancer Res. 1978;38:1345–1355. [PubMed] [Google Scholar]
- 20.Kobayashi S, Boggon TJ, Dayaram T, Jänne PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG, Halmos B. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 21.Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, Kris MG, Varmus H. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, Sawyers CL. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 23.Guix M, Faber AC, Wang SE, Olivares MG, Song Y, Qu S, Rinehart C, Seidel B, Yee D, Arteaga CL, Engelman JA. Acquired resistance to EGFR tyrosine kinase inhibitors in cancer cells is mediated by loss of IGF-binding proteins. J. Clin. Invest. 2008;118:2609–2619. doi: 10.1172/JCI34588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohren JF, Chen H, Pavlovsky A, Whitehead C, Zhang E, Kuffa P, Yan C, McConnell P, Spessard C, Banotai C, Mueller WT, Delaney A, Omer C, Sebolt-Leopold J, Dudley DT, Leung IK, Flamme C, Warmus J, Kaufman M, Barrett S, Tecle H, Hasemann CA. Structures of human MAP kinase kinase 1 (MEK1) and MEK2 describe novel noncompetitive kinase inhibition. Nat. Struct. Mol. Biol. 2004;11:1192–1197. doi: 10.1038/nsmb859. [DOI] [PubMed] [Google Scholar]
- 25.Lin H, Hestir K, Leo C, Huang M, Bosch E, Halenbeck R, Wu G, Zhou A, Behrens D, Hollenbaugh D, Linnemann T, Qin M, Wong J, Chu K, Doberstein SK, Williams LT. Discovery of a cytokine and its receptor by functional screening of the extracellular proteome. Science. 2008;320:807–811. doi: 10.1126/science.1154370. [DOI] [PubMed] [Google Scholar]
- 26.Hahn CK, Ross KN, Warrington IM, Mazitschek R, Kanegai CM, Wright RD, Kung AL, Golub TR, Stegmaier K. Expression-based screening identifies the combination of histone deacetylase inhibitors and retinoids for neuroblastoma differentiation. Proc. Natl. Acad. Sci. U.S.A. 2008;105:9751–9756. doi: 10.1073/pnas.0710413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gupta PB, Onder TT, Jiang G, Tao K, Kuperwasser C, Weinberg RA, Lander ES. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, Carpenter AE, Foo SY, Stewart SA, Stockwell BR, Hacohen N, Hahn WC, Lander ES, Sabatini DM, Root DE. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 29.Hao H, Muniz-Medina VM, Mehta H, Thomas NE, Khazak V, Der CJ, Shields JM. Context-dependent roles of mutant B-Raf signaling in melanoma and colorectal carcinoma cell growth. Mol. Cancer Ther. 2007;6:2220–2229. doi: 10.1158/1535-7163.MCT-06-0728. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.