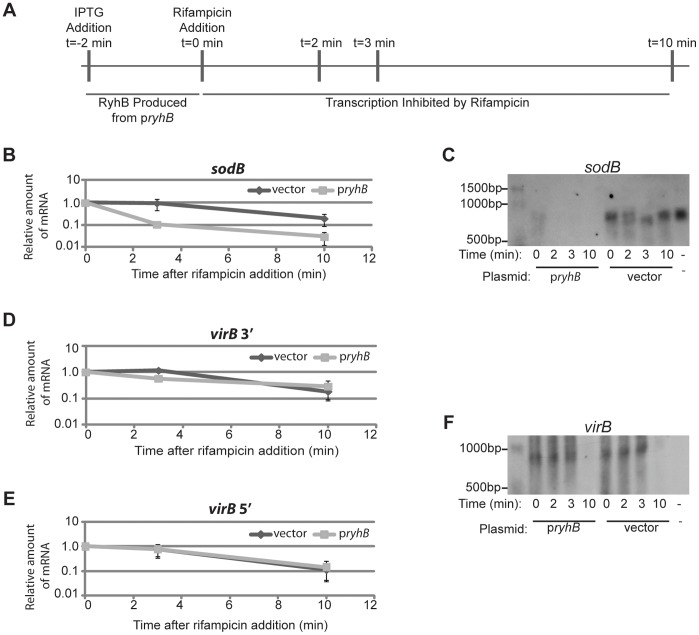
Figure 3. RyhB does not alter the stability of the virB mRNA molecule.
Schematic of the procedure used to induce RyhB production, inhibit transcription, and collect RNA samples for Real-time PCR and northern blot based investigations into the impact of RyhB on the stability of virB and sodB mRNA molecules (A). The relative amount of sodB mRNA (B) and virB mRNA (D and E) was quantified by Real-time PCR in the presence (light lines) and absence (dark lines) of increased RyhB production. At each time point the amount of mRNA present is expressed relative to the level of mRNA at the time of transcriptional inhibition by the addition of rifampicin (t=0) and is normalized to the amount of rrsA measured in the sample. The data is an average of three independent experiments and error bars represent one standard deviation. The same set of RNA samples was used to measure levels of both sodB and virB mRNA. Northern blot analysis was used to detect full-length sodB and virB mRNA in each RNA sample collected (C and F, respectively). The negative control lane in each northern blot contains RNA isolated from wild-type S. dysenteriae carrying the pQE vector grown at 30°C, as virB expression is inhibited at this temperature. Each northern blot shown is representative of analysis performed with three biological replicates.