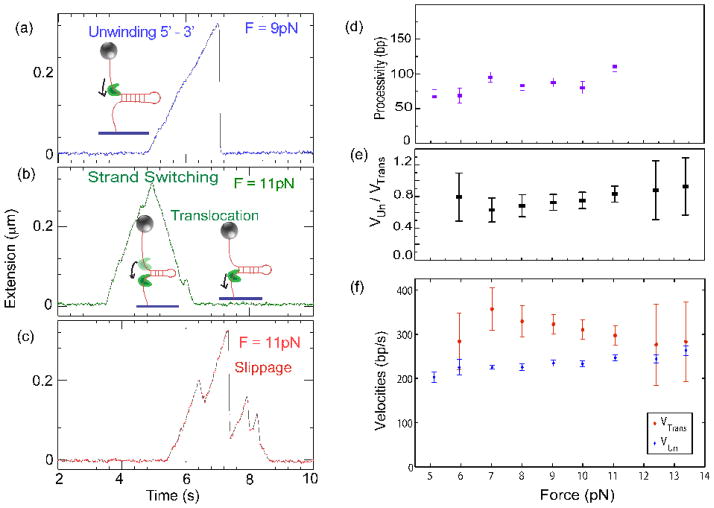
Fig. 8.
Single molecule unwinding and translocation. One end of a DNA hairpin substrate was attached to a glass coverslip, and the other end was attached to a magnetic bead. Magnets were used to pull the bead at a constant force. The force applied was low enough to prevent mechanical unzipping (below 15pN). This geometry enabled us to follow the enzyme dynamics as a function of the applied force. (a) A typical unwinding event presenting a regular speed of unwinding by Dda in the 5′-to-3′ direction, followed by enzyme dissociation and rapid re-annealing of the hairpin (at 9 pN, 1 base pair ~ 0.9nm). (b) Dda, during its course of unwinding, can sometimes switch strands and just translocate in the 5′-to-3′ direction. While the helicase is unwinding the 1200 base pair hairpin, its extension increases. Dda switches strands after unwinding about 300 base pairs (at 11 pN, 1 base pair unwound ~ 0.98 nm extension increase). The helicase then translocates and the hairpin rezips in its wake, at a rate limited by the translocation speed of the enzyme. Note that the rate of decrease in extension (translocation rate) is equivalent to the rate of increase of extension (unwinding rate). (c) An event demonstrating repeated unwinding due to slippages. In our experimental conditions, slippages occurred frequently. Dda processivity (d) unwinding rate (f), and translocation rate (f) versus applied force, measured in single molecule unwinding assays. (e) Shows the dependence on force of the ratio of the unwinding speed to the translocation speed. Notice that the translocation rate is obtained whenever Dda switches strands; as those events are short and sparse at low force, the error bars associated are rather large, and it was not possible to measure this parameter at 5pN.