Figure 1.
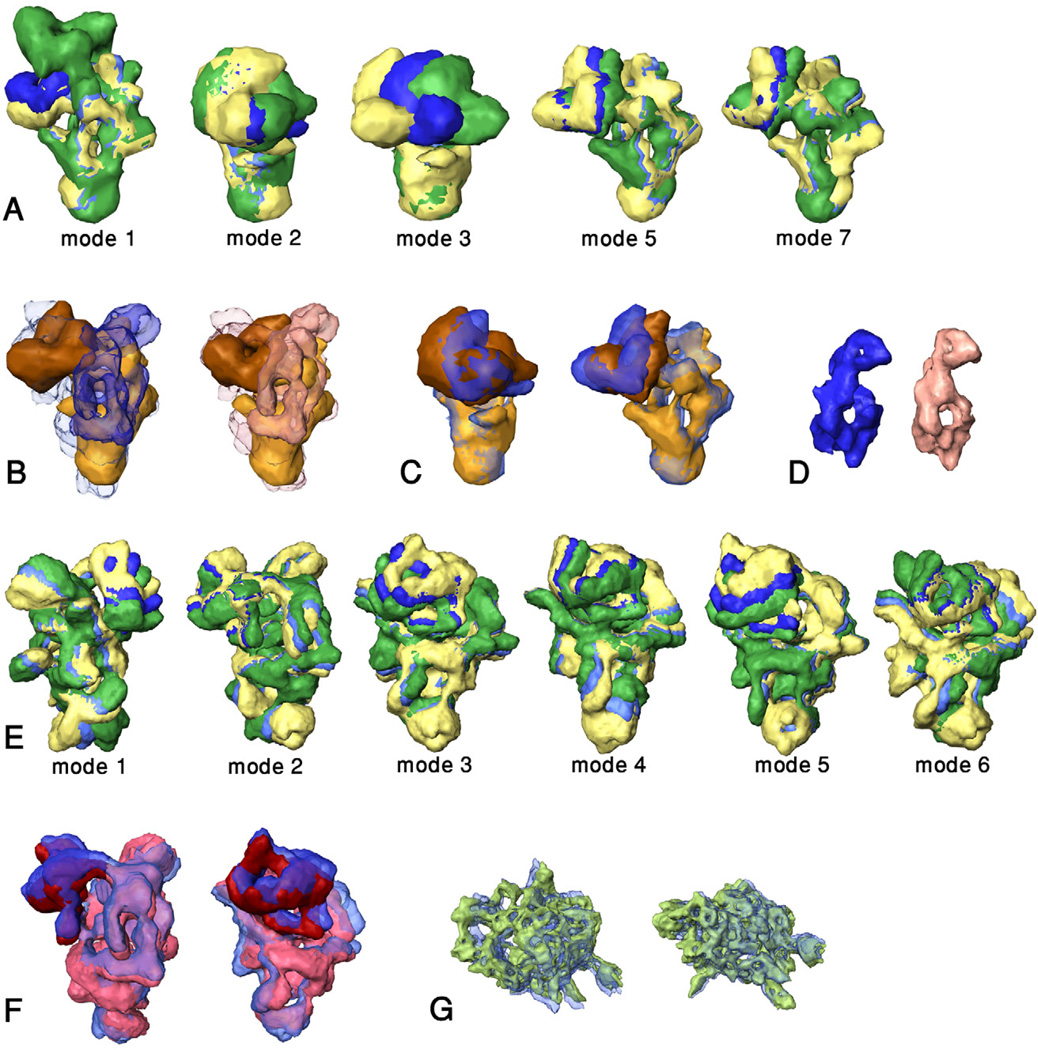
Matching ensembles of NMA conformers of spliceosomal subcomplexes into larger entities that contain them. Only normal modes that contribute to the best matching conformers are shown. The images are rotated to show the largest deformation. (A) Normal modes 1, 2, 3, 5 and 7 for U5 snRNP (the numbering excludes the six rigid body motion modes), that shape the conformers that match best within the free and “bound” U4/U6.U5 tri-snRNP. The unperturbed U5 snRNP is shown in blue. The yellow and green models depict the largest acceptable deformations (see Methods) in opposite directions. (B) The highest scoring match of U5 snRNP (gold body and brown head) within the unperturbed, “free” U4/U6.U5 tri-snRNP (blue; left image) and the “bound” U4/U6.U5 tri-snRNP (pink; right image). U4/U6.U5 tri-snRNP was made transparent in order to show the location of U5 snRNP. Note the empty blue or pink volume within the U4/U6.U5 tri-snRNP, which is attributed to the U4/U6 di-snRNP. (C) Left – comparison of the unperturbed U5 snRNP (transparent; blue with dark blue head) with the conformer that fits best within the free U4/U6.U5 tri-snRNP (gold with brown head). Right – similar comparison for the conformer that fits best within the “bound” U4/U6.U5 tri-snRNP. The structures were rotated to show the largest difference. (D) The U4/U6 di-snRNP density obtained by subtracting the density attributed to U5 snRNP from the density of the free or “bound” U4/U6.U5 tri-snRNP (blue and pink, respectively). (E) Normal modes 1 through 6 for U4/U6.U5 tri-snRNP; coloring as in A. (F) Two views comparing the unperturbed U4/U6.U5 tri-snRNP (transparent; dark blue head and blue body) with the best matching conformer of U4/U6.U5 tri-snRNP within the native spliceosome (dark red head and pink body). (G) Two views comparing the unperturbed SF3b complex (transparent; blue) with the conformer that fits best into native spliceosomeΔ[U4/U6.U5] (green). These images are not to scale with the other images.