Abstract
MHC class II molecules influence antigen-specific CD4+ T-lymphocyte responses primed by immunization and infection. CD4+ T-cell responses are important for controlling infection by many bacterial pathogens including Anaplasma marginale, and are observed in cattle immunized with the protective A. marginale outer membrane (OM) vaccine. Immunogenic proteins that comprise the protective OM vaccine include type IV secretion system (T4SS) proteins VirB9-1, VirB9-2, and VirB10, candidates for inclusion in a multi-epitope vaccine. Our goal was to determine the breadth of the VirB9-1, VirB9-2, and VirB10 T-cell response and MHC class II restriction elements in six cattle with different MHC class II haplotypes, defined by DRB3, DQA, and DQB allele combinations for each animal. Overlapping peptides spanning each T4SS protein were tested in T-cell proliferation assays with autologous antigen presenting cells (APC) and artificial APC expressing combinations of bovine DR and DQ molecules. Twenty immunostimulatory peptides were identified; three representing two or more epitopes in VirB9-1, ten representing eight or more epitopes in VirB9-2, and seven representing seven or more epitopes in VirB10. Of eight DRA/DRB3 molecules, four presented 15 peptides, which was biased as DRA/DRB3*1201 presented ten and DRA/DRB3*1101 presented four peptides. Four DQA/DQB molecules composed of two intrahaplotype and two interhaplotype pairs presented seven peptides, of which five were uniquely presented by DQ molecules. In addition,three functional mixed isotype (DQA/DRB3) restriction elements were identified. The immunogenicity and broad MHC class II presentation of multiple VirB9-1, VirB9-2, and VirB10 peptide epitopes justify their testing as a multi-epitope vaccine against A. marginale.
Introduction
Cell to cell interactions of the adaptive immune response are critically important for protection from pathogens. These interactions are orchestrated by the immunological synapse whose primary components are the T-cell receptor (TCR) on CD4+ T helper cells and major histocompatibility complex (MHC) class II molecules on antigen presenting cells (APC). The primary role of MHC class II molecules is to display peptides from exogenous proteins on the surface of the APC so they are available to interact with the TCR and initiate an antigen-specific CD4+ T-cell response (Germain 1986; Yewdell and Bennink 1990). MHC class II molecules comprise a set of peptide binding proteins of varying specificities (Unanue et al. 1989). In cattle, this diversity largely reflects the extensive allelic polymorphism used by the immune system to increase the antigen epitope-binding repertoire (Ellis and Ballingall 1999; Lewin et al. 1999; Park et al. 2004).
Cattle express two MHC class II proteins (DR and DQ), yet approximately one-half of haplotypes also have duplicated DQ regions (Glass et al. 2000; Norimine and Brown 2005). Mice express two class II proteins (H2-A and H2-E), and humans express three (HLA-DR, HLA-DQ, and HLA-DP). Genetic diversity of cattle is permitted through polymorphisms in DRB3, DQA, and DQB genes as well as through pairing of gene products, which can occur for any combination of α- and β-chains. In cattle, antigenic peptides are classically presented by monomorphic DRA paired with polymorphic DRB molecules to T cells (Brown et al. 2002; Glass et al. 2000; Norimine and Brown 2005; Norimine et al. 2006). However, antigenic peptides are also presented by intra- or interhaplotype pairs of DQA and DQB molecules (Brown et al. 2002; Glass et al. 2000; Moreno et al. 1990; Norimine and Brown 2005; Silk et al. 2005). Intrahaplotype pairing refers to the combination of DQA and DQB gene products encoded by alleles on the same chromosome (within the same haplotype). Interhaplotype pairing refers to the combination of DQA and DQB gene products encoded by alleles on different chromosomes (between different haplotypes). It has also been noted that isotype-mismatched combinations of DQ gene products with DR gene products, denoted mixed isotype, can be expressed with human HLA-DR and -DQ (Germain and Quill 1986; Lotteau et al. 1987) and murine H2-A and -E (Sant et al. 1987; Spencer et al. 1993) proteins. However, this has not been shown for cattle or any other species. If some alleles are more active than others for antigen presentation, protective immune responses against a bovine pathogen such as Anaplasma marginale will be strongly influenced by certain MHC class II alleles, and their characterization is therefore important for effective vaccine development.
The tick-borne pathogen A. marginale causes a persistent infection of cattle characterized by acute and chronic high-load bacteremia. Control measures against anaplasmosis are largely inadequate and the lack of a safe and effective vaccine results in large economic losses (Palmer et al. 2000). Protective immunity against disease is achieved by vaccination with a live, attenuated A. centrale vaccine, but this carries the risk of transmitting other blood-borne pathogens, and is not licensed for use in the United States. Protection against disease and, in some cases, infection can be achieved by immunization with purified A. marginale outer membranes (OM) (Tebele et al. 1991; Brown et al. 1998). However, the use of purified OM as a commercial vaccine is cost-prohibitive, so that identification of immunogenic and potentially protective proteins within the OM has been a focus of our research (Lopez et al. 2005; 2008). CD4+ T lymphocytes are important for inducing protective immune responses following immunization with OM (Brown et al. 1998). Effective vaccines against A. marginale will therefore likely require the inclusion of multiple conserved proteins or T-lymphocyte epitopes derived from these, because immunodominant, antigenically variant surface proteins such as major surface protein (MSP)2 have failed to elicit protective immunity (Abbott et al. 2005; Noh and Brown in press). Because MHC class II molecules influence antigen-specific CD4+ T-lymphocyte responses, characterization of MHC class II molecules and the identification of immunogenic T-cell epitopes are also important considerations for vaccine development.
Immunogenic proteins comprising the protective OM include the subdominant and conserved type IV secretion system (T4SS) proteins. The T4SS is a membrane protein complex of many bacterial pathogens, which secretes virulence factors and promotes host cell invasion and intracellular survival (Backert and Meyer 2006). It was previously shown that A. marginale T4SS proteins VirB9-1, VirB9-2, and VirB10 are strongly immunogenic for CD4+ T lymphocytes from A. marginale OM-immunized cattle and that these proteins are highly conserved across A. marginale strains and A. centrale (Lopez et al. 2007; Morse et al. 2012; Sutten et al. 2010). However, in a recent study, cattle 35160, 35280, and 35287, which are half-matched with respect to DRB3*1501, had differential responses to each protein (Morse et al. 2012). To understand the differences in T-cell responses observed for these animals to VirB9-1, VirB9-2, and VirB10 we sought to identify MHC class II-allele specific T-cell epitopes. Determining the breadth of the response to T-cell epitopes on VirB9-1, VirB9-2, and VirB10 in outbred cattle is important to develop a multiple-antigen and peptide-based vaccine construct.
To gain further insight into the antigenic structure of T4SS proteins VirB9-1, VirB9-2, and VirB10, we have investigated the proliferative responses of CD4+ T lymphocytes from MHC class II-defined cattle to a series of overlapping peptides spanning the length of each of these proteins. In addition, bovine cd80 and MHC class II DR- and DQ-transfected human embryonic kidney 293-F cells were used as artificial APC to determine MHC class II restriction elements. Our study highlights the importance of MHC class II DQ in addition to DR molecules in presenting pathogen-derived peptides to CD4+ T cells.
Materials and Methods
Antigens and synthetic peptides
Full-length protein sequences for VirB9-1, VirB9-2, and VirB10 of the St. Maries strain of A. marginale were used to synthesize peptides generally 30 amino acids in length and overlapping by ten amino acids. This yielded 13 peptides for VirB9-1, 14 peptides for VirB9-2, and 22 peptides for VirB10 (Table 1). The 30-mer VirB10 peptide 2 (P2) was difficult to synthesize, and was divided into two 20-mer peptides with 10 overlapping amino acids and re-named VirB10 P2.1 and P2.2, and an additional peptide representing the overlapping VirB10 P13 and P14 was also synthesized and designated P13-14. VirB9-1 P8-P13 and VirB10 P13-14 were synthesized and purified by Gerhard Munske, Laboratory for Biotechnology and Bioanalysis Unit 1, Washington State University (WSU). The remaining peptides were synthesized and purified by NeoPeptide (Cambridge, MA). All peptides were manufactured with 75% purity. Peptides were solubilized in 10% DMSO in PBS and diluted to a stock concentration of 1 mg/ml.
Table 1.
Sequences of overlapping peptides used to measure T-cell responses
| Protein | Peptide | Sequence |
|---|---|---|
| VirB9–1 | P1 | MKKAFMVCAVALLCSSAAFGKQEPRSIAAD |
| P2 | KQEPRSIAADDHIKIINFNPQSIHRYTGFY | |
| P3 | QSIHRYTGFYGYQSSILFESGEVIDTVSMG | |
| P4 | GEVIDTVSMGDSTGWQLVPKGNRLFIKPVG | |
| P5 | GNRLFIKPVGDNADTNVTIITNRRVYYFEL | |
| P6 | TNRRVYYFELHAEEASGLDDPRLAYEVRFV | |
| P7 | PRLAYEVRFVYPAASSVDAASSSDLGGGVS | |
| P8 | SSSDLGGGVSFPTYQNDVPDLSDPEVAKKG | |
| P9 | LSDPEVAKKGLNFDYSVSHTAGSANIVPIR | |
| P10 | AGSANIVPIRVFDDRKFTYMQFSNVNGDLP | |
| P11 | QFSNVNGDLPSIFNVDAEGYESLVNFRIVG | |
| P12 | ESLVNFRIVGDYVVVERVSPAFTLRYGSST | |
| P13 | AFTLRYGSSTACVFNEKLYRTSSTSRRGRG | |
| VirB9–2 | P1 | MNFYKNLLACSALLTVVFTGGVAQSAVSGG |
| P2 | GVAQSAVSGGAPVSVDSRIKTFVYSPNEIF | |
| P3 | TFVYSPNEIFTVVFNHGYHSFIEFSKGETI | |
| P4 | FIEFSKGETIKVMAMGDSVHWKVKPVDNKL | |
| P5 | WKVKPVDNKLFIMPLEREGKTNMLVETNKG | |
| P6 | TNMLVETNKGRSYAFDLVSKSAGPDAAGYK | |
| P7 | SAGPDAAGYKEVADELGRVDSPLLDMAYVV | |
| P8 | SPLLDMAYVVRFYYPDNNREFDLKGAGLAD | |
| P9 | FDLKGAGLADLSAPSLAKNPNSGEVTVRPN | |
| P10 | NSGEVTVRPNATGKNYVYSASSADATIVPV | |
| P11 | SSADATIVPVKTFDDGALTYFQFYDNNKVI | |
| P12 | FQFYDNNKVIPKVFSVGRHGKKVPCRMLLL | |
| P13 | KKVPCRMLLLKGYVIIEGVHKRLYLDYGKS | |
| P14 | KRLYLDYGKSGVEVVNTVL | |
| VirB10 | P1 | MSLGMSDETKDNNYGDGVEESVNVVGVHKS |
| P2.1 | SVNVVGVHKSKKLFVVLVVC | |
| P2.2 | KKLFVVLVVCAITGMAYYMF | |
| P3 | AITGMAYYMFFRGSGTTETSEEPQQVIEKQ | |
| P4 | EEPQQVIEKQDVDKLLKESEAPAQETAPRI | |
| P5 | APAQETAPRILTPPPKLPDLPPLVMPTAPE | |
| P6 | PPLVMPTAPELPTLARIAKKKKEEPVVEET | |
| P7 | KKEEPVVEETKEILPPAAESFFEPELQRRP | |
| P8 | FFEPELQRRPMEDDGPPQHIPMPYRPGGGA | |
| P9 | PMPYRPGGGAIPEPVPSFLGYDREKRGTPM | |
| P10 | YDREKRGTPMIVLGGGGDGGPSEDGGGQGT | |
| P11 | PSEDGGGQGTDSRFSTWSTLDGTSSPSVKA | |
| P12 | DGTSSPSVKATRVGDPGYVILQGHMIDAVL | |
| P13 | LQGHMIDAVLETAINSDIPGVLRAIVSRDV | |
| P13–14 | PGVLRAIVSRDVYAE | |
| P14 | VLRAIVSRDVYAEAGNMVMIPKGSRLIGSY | |
| P15 | PKGSRLIGSYFFDASGNNTRVTVSWSRVIL | |
| P16 | VTVSWSRVILPHGIDIQINSAGTDELGRNG | |
| P17 | AGTDELGRNGSAGFIDTKMGNVLTSTILLA | |
| P18 | NVLTSTILLAGVSMGTAFVTSKIPALQSEI | |
| P19 | SKIPALQSEIKDTTEEKGEKKKEEKSSTLP | |
| P20 | KKEEKSSTLPVKIVSDAVKDFSESMKALIK | |
| P21 | FSESMKALIKKYVDTSKPTIYVDQGTVMKV | |
| P22 | YVDQGTVMKVFVNQDIVFPREAVRR |
Cattle
Five age-matched steers and one cow (animal numbers: 35113, 35141, 35160, 35280, 35287, and 583) with varying and heterozygous MHC class II molecules were purchased from local dairies. Bovine lymphocyte antigen (BoLA) MHC class II DRB3 types were determined by the polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) method described previously (Morse et al. 2012; van Eijk et al. 1992) and DQA1, DQA2, DQB1, and DQB2 alleles were determined by sequencing entire cDNAs using previously described primer sets (Norimine and Brown 2005; Park et al. 2004). The class II haplotypes of all cattle are shown in Table 2. Cattle were immunized with A. marginale OM as previously described (Morse et al. 2012).
Table 2.
DRB3, DQA, and DQB alleles identified in the cattle used in this study
| Animal | Haplotype |
DRB3 |
DQA | DQB | |
|---|---|---|---|---|---|
| RFLP | Allele | Allele(s) | Allele(s) | ||
| 35113 | DH11A | 11 | *0902 | *0204 | *1803 |
| DH22H | 22 | *1101 | *1001 | *1002 | |
| *2206 | *1402 | ||||
| 35141 | DH22H | 22 | *1101 | *1001 | *1002 |
| *2206 | *1402 | ||||
| DH24A | 24 | *0101 | *0101 | *0101 | |
| 35160 | DH03A | 3 | *1001 | *1001 | *1002 |
| *2101 | *0901 | ||||
| DH16A | 16 | *1501 | *1001 | *1002 | |
| *2202 | *1301 | ||||
| 35280 | DH16A | 16 | *1501 | *1001 | *1002 |
| *2202 | *1301 | ||||
| DH27A | 27 | *1401 | *NDa | *1401 | |
| 35287 | DH16A | 16 | *1501 | *1001 | *1002 |
| *2202 | *1301 | ||||
| DH22H | 22 | *1101 | *1001 | *1002 | |
| *2206 | *1402 | ||||
| 583 | DH08A | 8 | *1201 | *1201 | *1005 |
| *2201 | *1201 | ||||
| DH23A | 23 | *2703 | *ND | *ND | |
| *ND | *ND | ||||
ND, not determined
Two week T-cell lines used for proliferation assays
Peripheral blood mononuclear cells (PBMC) from six A. marginale OM-immunized cattle were collected and depleted of CD8+ cells and γδ T cells using monoclonal antibodies (mAb) and complement lysis (Morse et al. 2012). Two-week cell lines from CD4+ T cell-enriched PBMC were stimulated with A. marginale OM as described (Morse et al. 2012). Proliferation assays were performed with 3 × 104 T cells and 2 × 105 irradiated autologous PBMC, as a source of APC, per well in complete RPMI-1640 medium (cRPMI, Brown et al. 1991) for 3-4 days. Positive control antigens included A. marginale OM and full-length recombinant VirB9-1, VirB9-2, and VirB10, expressed as described (Morse et al. 2012). These were used at a final concentration of 1 μg/ml and peptides were used at 0.1-20 μg/ml. The N-terminal peptide P1 from B. bovis rhoptry associated protein 1 (Rap1 P1) was used as a negative control (Norimine et al. 2002). Lymphocytes were radiolabeled during the last 18 h of culture with 3H-thymidine and the results are reported as stimulation index (SI). The SI was calculated by dividing the mean counts per minute (CPM) of replicate cultures with a test antigen by the mean cpm of cells cultured with medium. The SI for the different peptides were compared to the SI for Rap1 P1 using a one-way ANOVA corrected for multiple comparisons with the Dunnett’s test. Statistically significant T-cell stimulation by an antigen was set at P <0.05 and SI > 3.
Expression of BoLA-class II and CD80 molecules on artificial APC
Amplification of full-length cDNAs encoding sequences for DRA, DRB3, DQA, DQB, and CD80 was performed by PCR and each PCR product was cloned into the eukaryotic expression vector pCR3.1 (Invitrogen) as previously described (Norimine and Brown 2005). A mixture of 1 μg of each plasmid DNA encoding CD80, one class II α chain, and one class II β-chain, and 200 μl OptiMem (Gibco) plus 6 μl GeneJuice (Novagen) was incubated at room temperature for 10-15 min and then added to human embryonic kidney 293-F cells (Invitrogen) with 90% confluence on a six-well plate. Transfected 293-F cells were incubated and cultured for 2 days in a 37°C incubator as described (Norimine and Brown, 2005). Medium was removed and 50 μg/ml mitomycin C (Sigma-Aldrich) in cRPMI was added to each well and incubated for 2-2.5 h. The transfected cells were then harvested with HBSS containing 2 mM EDTA, washed, and used as APC with the OM-specific two week T-cell cell lines in proliferation assays.
Expression of BoLA-class II and CD80 molecules on transfected 293-F cells was verified by flow cytometry using 15 μg/ml bovine DR-specific mAb TH14B (IgG2a), DQ-specific mAb CC158 (IgG2a), and CD80 specific mAb IL-A159 (IgG1). TH14B was purchased from the WSU mAb center and mAbs CC158 and IL-A159 were kindly provided by the Institute of Animal Health, Compton, UK (Norimine and Brown 2005). For the secondary antibodies, R-phycoerythrin (R-PE)-conjugated goat anti-mouse IgG2a and R-PE-conjugated goat anti-mouse IgG1 (Caltag Laboratories) were used at a 1:200 dilution.
T-cell proliferation assays with transfected 293-F cells as APC
To determine DR and DQ molecules that present immunogenic peptides, transfected 293-F cells treated with mitomycin C were plated in 96-well round-bottomed plates at 5 × 104 cells/well, loaded with 20 μg/ml of an individual peptide in triplicate wells, incubated at 37°C for 1 h, washed three times, and used as APC with T cells from two week cell lines. Proliferation was measured after 3-4 days as previously described (Norimine and Brown 2005). For each peptide, T-cell proliferation to a peptide presented by 293-F cells transfected with CD80 and a pair of class II α and β chains was compared to proliferation using non-transfected 293-F cells, and significance was evaluated by the Student’s one-tailed t-test where P< 0.05.
Results
Breadth of the CD4+ T-cell response to VirB9-1, VirB9-2, and VirB10 peptides
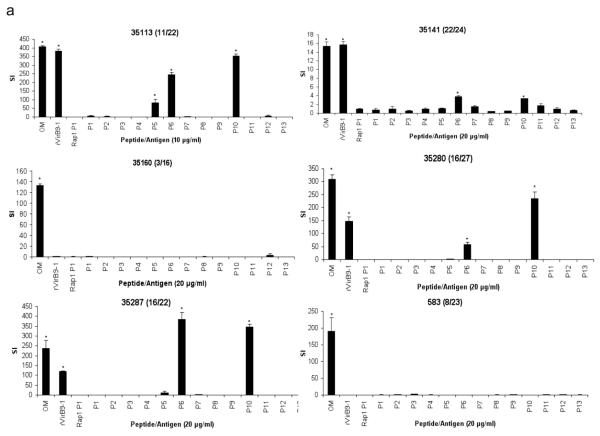
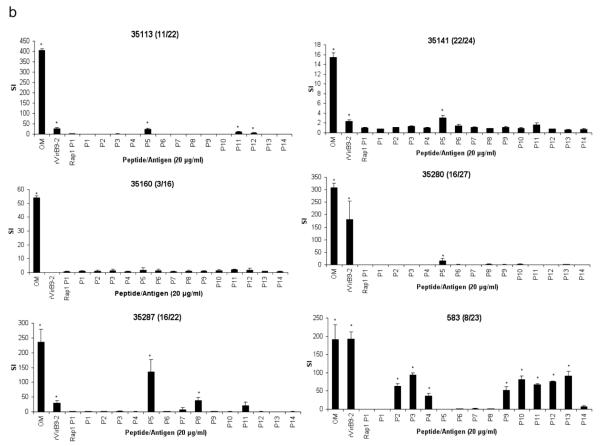
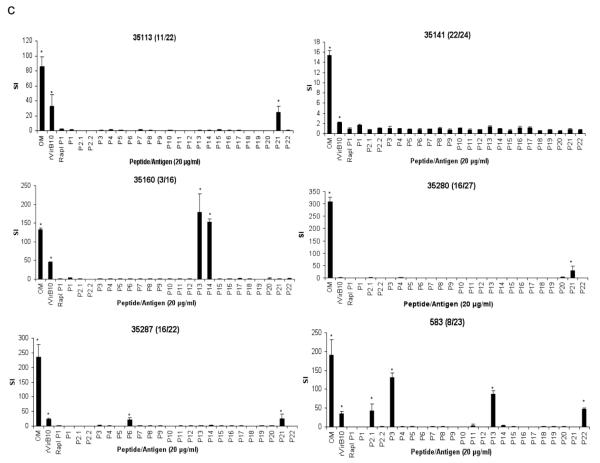
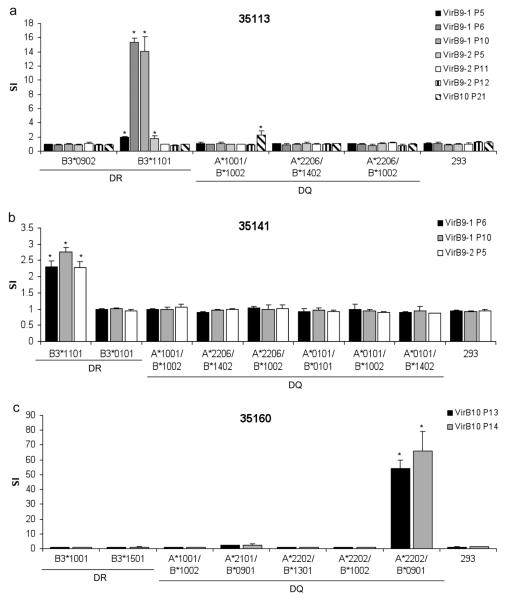
Holstein cattle expressing eight different DRB3 alleles were used determine the response to VirB9-1, VirB9-2, and VirB10 peptides. The DRB3 RFLP haplotypes and corresponding alleles (in parentheses) 3 (*1001), 8 (*1201), 11 (*0902), 16 (*1501), 22 (*1101), 23 (*2703), 24 (*0101), and 27 (*1401) are common among Holstein cattle. Of 878 Holstein calves from Washington and Vermont genotyped for DRB3 alleles in the past 10 years, the combined frequencies of these 8 alleles was 90% (see Online Resource 1). To assess the position and minimal number of T-cell epitopes, 30-mer peptides overlapping by 10 amino acids and spanning the complete sequences of VirB9-1, VirB9-2, and VirB10 were synthesized (Table 1). The peptides were tested for T-cell stimulation using OM-specific T-cell lines derived from animals 35113, 35141, 35160, 35280, 35287, and 583 with varying MHC class II haplotypes, specified in Table 2. VirB9-1 peptide-specific responses were detected with T-cell lines derived from animals 35113, 35141, 35280, and 35287, but not with T-cell lines from animals 35160 and 583, as the latter two cattle do not respond to full-length VirB9-1 (Fig. 1a). Animal 35113 (RFLP type 11/22) had significant T-cell responses against peptides VirB9-1 P5, P6, and P10. Animal 35287 (16/22) also responded to VirB9-1 P5, P6, and P10, although the response to P5 was not significant using a multiple comparison correction. T cells from animals 35141 (22/24) and 35280 (16/27) also recognized VirB9-1 P6 and P10.
Fig. 1.
Stimulation of CD4+ T-cell lines from A. marginale OM-immunized cattle with synthetic peptides from VirB9-1, VirB9-2, and VirB10. Short term T-cell lines from six animals (animal number and RFLP type in parenthesis is indicated on the top of each panel) were stimulated with 10 or 20 μg/ml of peptides from (a) VirB9-1 peptides P1-P13, (b) VirB9-2 peptides P1-P14, and (c) VirB10 peptides P1, P2.1, P2.2, and P3-P22. As a negative control, a 30-mer peptide derived from B. bovis Rap1 was used at an equal concentration. Additional controls were A. marginale OM and the full length recombinant protein used at 1 μg/ml. Results are presented as SI, and responses significantly higher than those for B. bovis Rap1 P1 are indicated with asterisks, where P <0.05
All cattle except 35160 (3/16), which did not respond to full-length VirB9-2, responded to one or more VirB9-2 peptides (Fig. 1b). VirB9-2 P5 was recognized by T cells from animals 35113 (11/22), 35141 (22/24), 35280 (16/27), and 35287 (16/22). Animal 35113 (11/22) also responded to VirB9-2 P11 and P12. Animal 35287 (16/22) also recognized VirB9-2 P8. Animal 583 (8/23) recognized eight peptides spanning VirB9-2, which were P2, P3, P4, P9, P10, P11, P12, and P13.
All cattle except 35141 responded to VirB10 peptides (Fig. 1c). Animal 35141 weakly responded to recombinant VirB10, however no responses to the peptides were observed. VirB10 P21 was recognized by T cells from cattle 35113 (11/22), 35280 (16/27), and 35287 (16/22). It was previously reported that animal 35280 did not recognize recombinant VirB10 (Morse et al. 2012), but the response to VirB10 is inconsistent for this animal, as we found significant proliferation to VirB10 in two of six assays (Fig. 1c and data not shown). In one of those assays where the response was positive, significant T-cell proliferation to VirB10 P21 by this animal was found. Animal 35287 (16/22) also recognized VirB10 P6. T-cell lines derived from animal 35160, which responded only to VirB10, recognized VirB10 P13 and P14. T cells from animal 583 (8/23) recognized VirB10 P2.1, P3, P13, and P22.
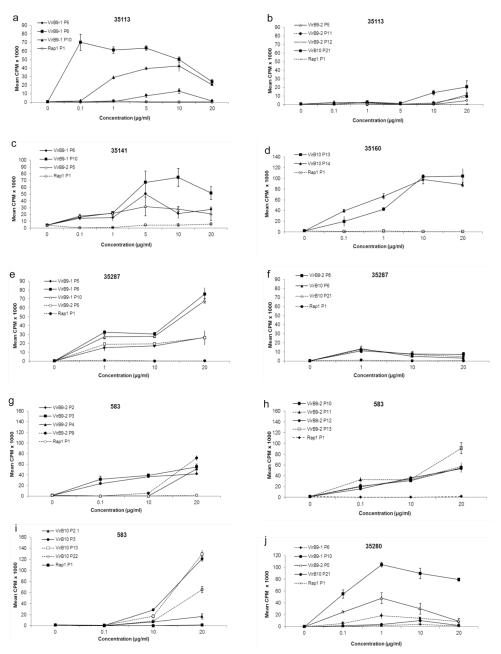
Dose response curves shown in Fig. 2 yielded interesting information about the strength of the T-cell response for specific peptides from each animal. For animals 35113 (11/22), 35141 (22/24), and 35287 (16/22) the strongest T-cell responses were induced by VirB9-1 P6 and VirB9-1 P10, which were significant at all the concentrations tested (Fig. 2a,c,e). Likewise, animal 35280 (16/27) also had a strong T-cell response to VirB9-1 P10 and the next strongest response from this animal was induced by VirB9-2 P5 (Fig. 2j). For animal 35160 (3/16), VirB10 P13 and P14 induced similar T-cell responses and these were still significant at 0.1 μg/ml peptide (Fig. 2d). The fact that these peptides overlap in sequence by ten amino acids suggested the presence of a single epitope in the overlapping region. VirB9-2 P2, P3 and P10-P13 induced the strongest T-cell responses from 583 (8/23), that were significant using as little as 0.1 μg/ml peptide, which represent a minimum of one and three epitopes, respectively (Fig. 2g,h). There was not a single peptide that induced T-cell responses from every animal, yet all four animals that responded to VirB9-1 had strong T-cell responses to P10. Induction of strong T-cell responses at lower peptide concentrations indicates a relatively higher avidity interaction, and peptides with this characteristic would be best for inclusion in a vaccine. For all six MHC class II-diverse animals the peptides with highest avidity were VirB9-1 P6 and P10, VirB9-2 P2, P3, P5 P10, P11, P12, and P13, and VirB10 P13 and P14. Further mapping studies will need to be performed to define the minimal amino acid sequences representing epitopes in these 30-mer peptides, which typically range from 9-12 amino acids.
Fig. 2.
Dose-dependent proliferative responses to stimulatory peptides. Short-term T-cell lines from six animals (a) 35113, (b) 35113, (c) 35141, (d) 35160, (e) 35287, (f) 35287, (g) 583, (h) 583, (i) 583, and (j) 35280, were stimulated with the 0.1 to 20 μg/ml of the indicated peptides and cultured for three days, radiolabeled, and harvested. As a negative control, B. bovis Rap1 P1 was used at an equal concentration. Proliferation is presented as the mean CPM
DR and DQ expression and antigen presentation
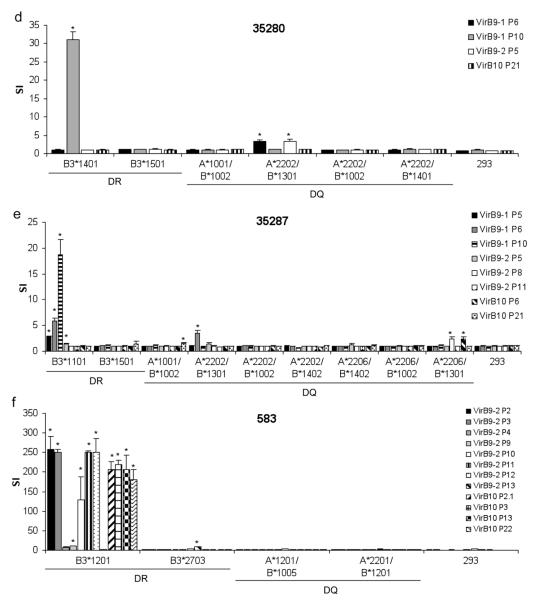
To assess the peptide presentation by MHC class II molecules for each animal, artificial APC were loaded with individual peptides, and used in T-cell assays. Artificial APC were generated by transfecting 293-F cells with all possible combinations of DRA and DRB3, DQA and DQB, DRA and DQB, and DQA and DRB3 alleles known for each animal. However, not all of the possible DR and DQ combinations available for each animal were expressed on the surface of transfected 293-F cells. The combinations that did express are listed in Table 3. Each of the expressed MHC class II combinations was tested for the ability to present peptides to T cells from cattle expressing the corresponding alleles (Fig. 3). T cells from animals 35113, 35141, and 35287 were stimulated with DRA/DRB3*1101 presenting peptides VirB9-1 P6, P10, and VirB9-2 P5 (Fig. 3a,b,e). Also, DRA/DRB3*1101 presented VirB9-1 P5 to T cells from animals 35113 and 35287 (Fig. 3a,e). T cells from 35280 also responded to VirB9-1 P6, P10, and VirB9-2 P5, but unlike animals 35113, 35141, and 35287, 35280 does not have a DRA/DRB3*1101 combination (Table 2). The class II molecules presenting these peptides to T cells derived from animal 35280 were identified as DRA/DRB3*1401 for VirB9-1 P10, and DQA*2202/DQB*1301 for VirB9-1 P6 and VirB9-2 P5 (Fig. 3d). Thus, VirB9-1 P6 and P10, and VirB9-2 P5 were presented by multiple MHC class II molecules. The presentation of VirB9-1 P6 by DQA*2202/DQB*1301 was also observed using T cells from animal 35287 (Fig. 3e). VirB10 P21 was presented by DQA*1001/DQB*1002 to T cells from animals 35113 and 35287 (Fig. 3a,e). Animals 35141, 35160, and 35280 also had this DQ combination, but significant responses to VirB10 P21 were not detected using this DQ pair (Fig. 3b,c,d). For animal 35113, we were unable to identify a restriction element for VirB9-2 P11 and P12 (Fig. 3a), possibly because responses to these peptides were relatively weak, and only detected with 20 μg/ml peptide (Fig. 2b).
Table 3.
Expressed BoLA class II α/β combinations
| BoLA class II molecule | Percentage of cells expressing class II |
||
|---|---|---|---|
| Isotype | A chain | B chain | |
| DRa | |||
| *0101 | *0101 | 49.6 | |
| *0902 | 56.5 | ||
| *1001 | 53.0 | ||
| *1101 | 61.8 | ||
| *1201 | 68.0 | ||
| *1401 | 60.8 | ||
| *1501 | 51.7 | ||
| *2703 | 60.0 | ||
| DQb-Intrahaplotype | |||
| *0101 | *0101 | 34.5 | |
| *1001 | *1002 | 59.7 | |
| *1201 | *1005 | 45.7 | |
| *2101 | *0901 | 42.0 | |
| *2201 | *1201 | 49.5 | |
| *2202 | *1301 | 52.4 | |
| *1002 | 30.6 | ||
| *2206 | *1402 | 59.9 | |
| *1002 | 37.1 | ||
| Interhaplotype | |||
| *0101 | *1002 | 45.6 | |
| *1402 | 56.8 | ||
| *2202 | *0901 | 57.5 | |
| *1401 | 16.8 | ||
| *1402 | 57.0 | ||
| *2206 | *1301 | 17.6 | |
| Mixed Isotypea,c | |||
| *0101 | *0101 | 60.0 | |
| *1001 | *0101 | 55.7 | |
| *1001 | 39.4 | ||
| *1501 | 29.2 | ||
| *1201 | *1201 | 44.8 | |
| *2101 | *1001 | 22.2 | |
| *1501 | 33.4 | ||
| *2201 | *1201 | 31.4 | |
| *2202 | *1001 | 38.7 | |
| *1501 | 40.0 | ||
| *1401 | 19.4 | ||
| *1101 | 23.9 | ||
| *2206 | *0101 | 52.4 | |
| *1101 | 44.3 | ||
| *1501 | 33.0 | ||
Expression of all combinations containing DR was detected using Th14B mAb.
Expression of all intra- and interhaplotype DQ combinations was detected using CC158 mAb.
Combinations of DRA and DQB were not detected, and only DQA and DRB combinations that were detected are shown.
Fig. 3.
Identification of BoLA class II restriction elements for VirB9-1, VirB9-2, and VirB10 epitopes using 293-F cells expressing bovine DR and DQ molecules. Short-term T-cell lines from six animals (a) 35113, (b) 35141, (c) 35160, (d) 35280, (e) 35287, and (f) 583 were cultured for three days with human embryonic kidney 293-F cells expressing the indicated CD80 and DR or DQ molecules that were loaded with 10 μg/ml of the indicated peptide. Results are presented as SI and responses T-cell responses using each artificial APC pulsed with an individual peptide were compared to those using non-transfected 293-F cells incubated with the same peptide. Significant responses were determined using a Student’s one-tailed t-test, where P < 0.05, and are indicated by asterisks
T cells from animal 35160 did not respond to VirB9-1 or VirB9-2 peptides, and responded only to P13 and P14 from VirB10, which were presented by DQA*2202/DQB*0901 (Fig. 3c). Because the responses to these two overlapping peptides were comparable and both peptides were presented by the same class II molecule, we also tested a 15-mer peptide P13-14 (Table 1) that contains the overlapping 10-mer sequence plus two and three amino acids on the N- and C-termini, respectively, for presentation by artificial APC. The responses to P13, P14, and P13-14 were comparable and all were presented by both irradiated autologous PBMC and DQA*2202/DQB*0901 transfected artificial APC (data not shown), suggesting a single epitope, VLRAIVSRDV, is recognized. This DQA/DQB combination was clearly an interhaplotype DQ molecule since the DQA*2202 allele is associated with the DH16A haplotype and the DQB*0901 allele is associated with the DH03A haplotype (Table 2). Therefore, in order to express DQA*2202/DQB*0901, a bovine must be heterozygous. Another interhaplotype combination available only for 35287 was DQA*2206/DQB*1301, which presented VirB9-2 P8 and VirB10 P6 (Fig. 3e). This result explains why 35287 was the only animal to respond significantly to these peptides.
As compared to the other five animals, animal 583 has unique DR and DQ combinations (Table 2), and different responses to VirB9-2 and VirB10 peptides were observed. DRA/DRB3*1201 presented the majority of the peptides to T cells from animal 583: VirB9-2 P2, P3, P9, P10, P11, and P12, as well as VirB10 P2.1, P3, P13, and P22 (Fig. 3f). We were unable to identify a restriction element for VirB9-2 P4, possibly because the response to this peptide was relatively weak, and only detected with 20 μg/ml peptide (Fig. 2g). VirB9-2 P13 was presented by DRA/DRB3*2703 (Fig. 3f).
MHC class II molecules shown to present immunostimulatory VirB9-1, VirB9-2, and VirB10 peptides to CD4+ T cells are summarized in Table 4. We identified four DRA/DRB3 molecules that presented the majority of peptides. Two intrahaplotype DQ combinations presented three peptides: DQA*2202/DQB*1301 presented VirB9-1 P6 and VirB9-2 P5 and DQA*1001/DQB*1002 presented VirB10 P21. We also identified two interhaplotype DQ combinations that presented four peptides: DQA*2206/DQB*1301 presented VirB9-2 P8 and VirB10 P6 and DQA*2202/DQB*0901 presented VirB10 P13 and P14 (and the overlapping P13-14). Five of these peptides were not presented by DRA/DRB3 molecules. The MHC class II DQ interhaplotype pair DQA*2202/DQB*0901 was the only functional class II molecule for animal 35160, and it presented the only T-cell epitope(s) identified on VirB10. Thus, interhaplotype pairing was extremely important in this animal for recognition of VirB10.
Table 4.
Summary of BoLA-class II restriction elements presenting peptides to T cells
| BoLA-class II restriction element |
Presented peptide | Animal(s) # | |
|---|---|---|---|
| Isotype | Allele | ||
| DR | |||
| DRA/DRB3*1101 | VirB9-1 P5 | 35113, 35287 | |
| VirB9-1 P6 | 35113, 35141, 35287 | ||
| VirB9-1 P10 | 35113, 35141, 35287 | ||
| VirB9-2 P5 | 35113, 35141, 35287 | ||
| DRA/DRB3*1401 | VirB9-1 P10 | 35280 | |
| DRA/DRB3*1201 | VirB9-2 P2 | 583 | |
| VirB9-2 P3 | 583 | ||
| VirB9-2 P9 | 583 | ||
| VirB9-2 P10 | 583 | ||
| VirB9-2 P11 | 583 | ||
| VirB9-2 P12 | 583 | ||
| VirB10 P2.1 | 583 | ||
| VirB10 P3 | 583 | ||
| VirB10 P13 | 583 | ||
| VirB10 P22 | 583 | ||
| DRA/DRB3*2703 | VirB9-2 P13 | 583 | |
| DQ | |||
| Intrahaplotype | |||
| DQA*1001/DQB*1002 | VirB10 P21 | 35113, 35287 | |
| DQA*2202/DQB*1301 | VirB9-1 P6 | 35280, 35287 | |
| VirB9-2 P5 | 35280 | ||
| Interhaplotype | |||
| DQA*2202/DQB*0901 | VirB10 P13 | 35160 | |
| VirB10 P14 | 35160 | ||
| DQA*2206/DQB*1301 | VirB9-2 P8 | 35287 | |
| VirB10 P6 | 35287 | ||
| Mixed isotype | |||
| DQA*2206/DRB3*1101 | VirB9-2 P5 | 35113, 35141, 35287 | |
| DQA*1201/DRB3*1201 | VirB9-2 P12 | 583 | |
| DQA*2201/DRB3*1201 | VirB9-2 P12 | 583 | |
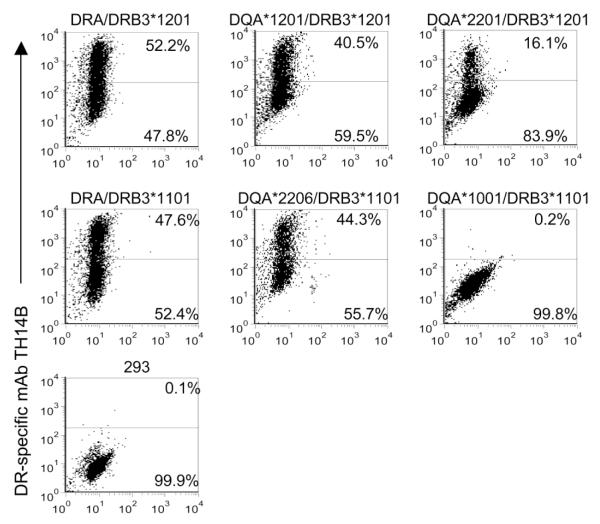
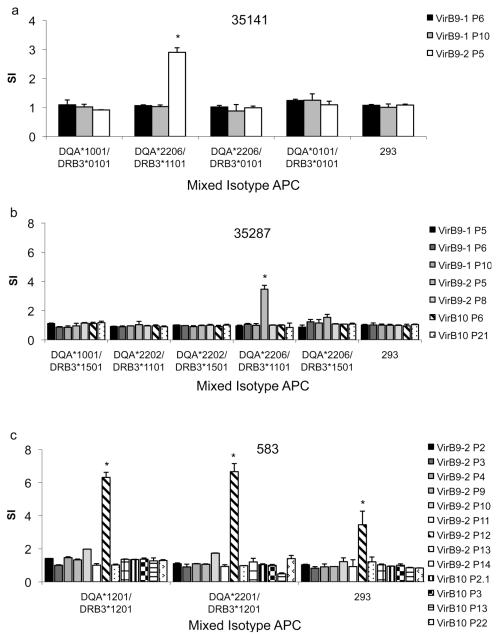
Mixed isotype pairs containing DQA and DRB3 alleles were expressed on the transfected 293-F cell surface, so their abilities to present peptides for each animal were also examined. Each combination that was expressed, listed in Table 3, was tested with T cells from animals that had the corresponding alleles. We determined that mixed isotypes DQA*1201/DRB*1201, DQA*2201/DRB3*1201, and DQA*2206/DRB3*1101 presented peptides. Expression of these mixed isotype class II molecules, as determined by flow cytometry analysis using mAb TH14B, is shown in Fig. 4. Fig. 4 also includes the positive controls DRA/DRB3*1201 and DRA/DRB3*1101 and negative controls DQA*1001/DRB3*1101 (which did not express) and non-transfected 293-F cells. VirB9-2 P5 was presented by DQA*2206/DRB3*1101 to T cells derived from cattle 35141 and 35287 (Fig. 5a,b). T-cell lines from animal 583 responded to the mixed isotype combinations DQA*1201/DRB*1201 and DQA*2201/DRB3*1201 presenting VirB9-2 P12 (Fig. 5c). However, a weak T-cell response to this peptide was also still present when non-transfected 293-F cells were used. This can be explained by residual peptide remaining in the well despite repeated washing, and residual autologous APC in the two week cell lines from this animal, but the responses to VirB9-2 P12 presented by DQA*1201/DRB*1201 and DQA*2201/DRB3*1201 were nevertheless significantly greater than the response with 293-F cells alone.
Fig. 4.
Expression of peptide-presenting mixed isotype pairs. Human embryonic kidney 293-F cells expressing CD80 and the indicated mixed isotype BoLA class II molecules were stained with DR-specific mAb TH14B and secondary antibody R-PE conjugated goat anti-mouse IgG2a, and analyzed by flow cytometry. Positive controls were mAb TH14B labeled DRA/DRB3 molecules and negative controls were mAb TH14B stained non-transfected 293-F cells and DQA*1001/DRB3*1101, which failed to express
Fig. 5.
Identification of BoLA class II restriction elements for VirB9-1, VirB9-2, and VirB10 epitopes using 293-F cells expressing bovine class II mixed isotypes. Short term T-cell lines from six animals (a) 35141, (b) 35287), and (c) 583 were cultured for three days with human embryonic kidney 293-F cells expressing CD80 and the indicated mixed isotype BoLA class II molecules that were loaded with 10 μg/ml of the indicated peptide. Results are presented as SI and T-cell responses using artificial APC pulsed with an individual peptide were compared to those of non-transfected 293-F cells incubated with the same peptide. Significant responses were determined using a Student’s one-tailed t-test, where P < 0.05, and are indicated by asterisks
Discussion
A. marginale OM can induce complete protection against disease, and in some animals, infection (Brown et al. 1998; Tebele et al. 1991, Noh and Brown, in press). However, it is unfeasible to use purified OM as a commercial vaccine because the bacteria are difficult to grow in culture and need to be harvested from infected cattle blood. Using blood-derived bacteria poses risks including potential contamination with other blood-borne pathogens and erythrocyte membranes. Use of individual immunodominant surface proteins, such as MSP2, has failed to provide protection against disease (Abbott et al. 2005, Noh and Brown in press). For these reasons, vaccine development for anaplasmosis has focused on identifying subdominant but immunogenic, conserved proteins in the OM (Lopez et al. 2005, 2008). Furthermore, we have evidence that association of proteins within the OM is important for providing increased T-cell help for IgG production (Macmillan et al. 2008; Morse et al. 2012). Therefore a protective vaccine is likely to require multiple subdominant proteins or combinations of individual linked T- and B-cell epitopes derived from the same or associated OM proteins. It would be advantageous to use multiple epitopes derived from multiple proteins rather than multiple proteins if the vaccine were to be delivered by a viral vector. There are limitations on the amount of cDNA that can be expressed as protein in these types of vectors.
A. marginale T4SS proteins VirB9-1, VirB9-2, and VirB10 are strongly immunogenic for CD4+ T lymphocytes from OM-immunized cattle (Lopez et al., 2007; Sutten et al., 2010; Morse et al., 2012). Animals 35113, 35141, and 35287, which are half-matched with respect to DRB3*1101 and associated DQ molecules, recognized all three of these proteins. However, animals 35160, 35280, and 35287, which are half-matched with respect to DRB3*1501 and associated DQ molecules, had differential responses, in that 35160 did not respond to VirB9-1 or VirB9-2, and 35280 responded weakly and inconsistently to VirB10. To understand the differences in T-cell responses observed for these animals to VirB9-1, VirB9-2, and VirB10 we sought to identify MHC class II-allele specific T-cell responses to overlapping peptides. The differential T-cell responses were in part explained by presentation of peptides by other class II molecules not shared between animals or by unique combinations of DQA and DQB molecules. However, when the alleles encoding functional class II molecules were present, CD4+ T-cell lines from these three animals did not always respond to the same peptides, as observed for VirB9-1 P6, VirB9-2 P5, and VirB10 P21. VirB9-1 P6 and VirB9-2 P5 were presented by DQA*2202/DQB*1301, an intrahaplotype pair encoded by alleles present in all three cattle, yet neither peptide was recognized by animal 35160. Similarly, VirB10 P21 was presented by intrahaplotype pair DQA*1001/DQB*1002, also encoded by alleles found in all three animals but not recognized by 35160. It is possible that not all class II molecules expressed in 293-F cells are expressed and functional in each individual animal, and haplotype combinations may affect preferential pairing of alpha and beta molecules in vivo. Since expressed BoLA class II molecules are comprised of many functional intra- and interhaplotype DQ molecules and potentially mixed isotypes, the population of antigen-specific CD4+ T cells could also differ qualitatively and quantitatively among individual animals.
In summarizing the information for all cattle in this study, 20 immunostimulatory peptides and 11 peptide-presenting elements were identified. These included four DRA/DRB3 pairs, two intrahaplotype-DQA/DQB pairs, two interhaplotype-DQA/DQB pairs, and three DQA/DRB3 mixed isotype pairs. Thus, consistent with previous studies, it was very important to identify DQ-restriction elements that uniquely present pathogen peptide epitopes to CD4+ T cells (Brown et al. 2002; Glass et al. 2000; Norimine and Brown 2005). Notably, we now understand the differential T-cell responses observed for these cattle, and can predict T-cell responses to these T4SS proteins and epitopes by cattle with these defined haplotypes. Furthermore, the identification of T-cell epitopes on VirB9-1, VirB9-2, and VirB10 can be used to develop a multiple-antigen, epitope-based vaccine construct.
The four DRA/DRB3 combinations presenting peptides were DRA/DRB3*1101, DRA/DRB3*1401, DRA/DRB3*1201, and DRA/DRB3*2703. The function of DRA/DRB3*1201 in presenting peptides to T cells from animal 583 was important, presenting a minimum of four epitopes from VirB9-2 and four from VirB10, when considering that contiguous overlapping peptides contain a minimum of one epitope. BoLA DRB3 genes have been associated with resistance and susceptibility to several viral, bacterial, and parasitic infections, including bovine leukemia virus infection, (Amills et al. 1998; Xu et al. 1993; Zanotti et al. 1996), foot and mouth disease (FMD) (Alizadeh et al. 2003; Haghparast et al. 2000), Staphylococcus aureus-associated mastitis (Park et al. 2004; Rupp et al. 2007), neosporosis (Schwab et al. 2009), and East Coast fever (Ballingall et al. 2004). However, despite the indication of the role that BoLA-DR molecules play in protective immunity, very few studies have performed functional assays with BoLA class II molecules. Among these studies pathogen epitope restriction elements have been identified for DRA/DRB3*1201 presenting A. marginale peptides MSP2 P16-7 (Norimine and Brown 2005) as well as MSP1a peptide F2-1-1b (Norimine et al. 2006). DRB3*1101 presented MSP1a F2-B to T cells (Norimine and Brown 2005). Several FMD viral peptides were shown to be presented by DRA/DRB3*1101, DRA/DRB3*1201, and DRA/DRB3*2703 (Alizadeh et al. 2003; Haghparast et al. 2000).
In cattle, duplication of DQ molecules amplifies the opportunity for intra- and interhaplotype pairing to form functional heterodimers, thereby increasing the complexity of restriction elements and providing more dynamic immune responses (Glass et al. 2000; Norimine and Brown 2005). We identified the products of two interhaplotype DQ molecules, DQA*2202/DQB*0901 and DQA*2206/DQB*1301. Here, these presented peptides from VirB9-2 and VirB10. This is the first study to report a functional DQA*2202/DQB*0901 molecule, which presented VirB10 P13, P14 and overlapping P13-14. This DQ combination was the only antigen-presenting element identified for animal 35160, explaining its T-cell response to VirB10. Interhaplotype DQA*2206/DQB*1301 was previously reported to present A. marginale MSP1a peptide B to T cells (Norimine and Brown 2005) and is apparently not rare because cattle heterozygous for the RFLP type 16/22 are common in Washington State Holstein herds (unpublished observations). The α- and β-chains of this combination may also have some flexibility as to which protein they partner with (Tables 3 and 4 and ref. Norimine and Brown 2005). Furthermore, even though not all possible DQ combinations were expressed on the cell surface, we have observed that there is preferential pairing of DQ molecules and that several DQ molecules observed in this study were noted in previous studies from our lab using different cattle and antigens (Brown et al. 2002; Norimine and Brown 2005; Norimine et al. 2006).
Three mixed isotype combinations that presented peptides were also identified, i.e. DQA*2206/DRB3*1101 presented VirB9-2 P5, and DQA*1201/DRB3*1201 and DQA*2201/DRB3*1201 presented VirB9-2 P12. However, the corresponding DRA/DRB3 combination for these mixed isotypes also presented the same peptide. This suggests that the β-chain may be more important for presenting the two peptides and the α-chain is interchangeable. This is the first documentation that bovine mixed isotypes have the capability to present peptides; however there is precedence for this from studies with mice and humans (Germain and Quill 1986; Lotteau et al. 1987; Sant et al. 1987; Spencer et al. 1993). Despite mixed isotype combinations being a relatively unexplored area, there remains the possibility that this represents a valid mechanism for antigen presentation, but further analyses are required to determine how important this is in vivo. We cannot rule out that the bovine mixed isotype peptide presentation is an in vitro artifact, as mAb that detect mixed isotypes are not available to confirm that this presentation occurs in vivo.
Selecting peptides that contain CD4+ T-cell epitopes with high avidity TCR interactions is an essential step for the construction of a multiple-antigen and peptide-based vaccine. Also, vaccine epitopes should be recognized broadly by diverse MHC class II molecules expressed in the target population. For A. marginale T4SS proteins, the peptides that have the highest avidity, based on stimulating T-cell proliferation at a low concentration, and are recognized by common MHC class II molecules in cattle are VirB9-1 P6 and P10, VirB9-2 P2, P3, P5, P10, P11, and P12, and VirB10 P13 and P14. VirB9-1, VirB9-2, and VirB10 sequences are highly conserved across A. marginale strains and A. centrale (Lopez et al., 2007; Morse et al. 2012), and sequences of peptides with the highest avidity (VirB9-1 P5, VirB9-2 P2, P5, P12, and P13, and VirB10 P13 and P14) are completely conserved (100% identical). The peptide sequences for VirB9-1 P10 and VirB9-2 P3 are also completely conserved among A. marginale strains and have only one amino acid difference in A. centrale. Similarly, VirB9-2 P10 and P11 are completely conserved among all strains of A. marginale, but have four and three amino acid differences, respectively, in A. centrale. Because both A. marginale OM and live A. centrale provide protective immunity against disease following A. marginale infection, this high degree of conservation in peptide epitopes between these bacteria indicates they may contribute to the protective immune response.
The identification of MHC class II restricted antigenic peptides for inclusion in a vaccine is a high research priority. However, bioinformatic tools have not been developed to accurately predict whether a given peptide will interact with BoLA class II molecules (Jones et al. 2011; Lafuente and Reche 2009). The prediction accuracy is dependent on the experimental datasets used, and for the bovine system this is severely lacking. In this study, we used six cattle with different haplotypes to identify the MHC class II restriction elements for peptides spanning three immunogenic T4SS proteins. Upon identification of minimal epitopes, the results will be useful to help formulate a prediction model for peptide binding to bovine MHC class II DR, and importantly, DQ molecules.
Different combinations of heterozygous MHC class II molecules in cattle increases the number of pathogen-derived peptide epitopes presented to T cells. We have identified highly antigenic CD4+ T-cell epitopes on T4SS proteins VirB9-1, VirB9-2, and VirB10 presented by BoLA class II DR and DQ molecules that are encoded by alleles prevalent among Holstein cattle worldwide. Similar frequencies of the MHC class II alleles studied here were found for Holstein cattle in Japan, where the combined frequencies were 81.2%, 86.9%, 90.3%, and 95.2% in four populations studied (Miyasaka et al., 2011; Takeshima et al., 2003), in Ontario, Canada, where the combined frequency was 89.5% (Sharif et al., 1998), in Iran, where the combined frequencies were 73.2%, 75.5%, 78.1%, and 80.2% in several studies (Nassiry et al., 2008; Pashimi et al., 2009), and in Europe, where the combined frequency was 89.5% (Nassiry et al., 2008). Inclusion of VirB9-1, VirB9-2, and VirB10 peptides linked together in a multiple-antigen and peptide-based vaccine will not only provide more than one immunogenic peptide, but will offer the opportunity for increased T-cell dependent IgG responses upon infection if B cell epitopes are also included (Morse et al., 2012). Furthermore, if protective, such a vaccine would be useful to protect the majority of individuals within Holstein populations across the globe. Thus, a vaccine made up of highly conserved VirB9-1, VirB9-2, and VirB10 peptides will be tested in a future study to protect cattle against A. marginale.
Supplementary Material
Acknowledgments
The technical assistance of Shelley Whidbee, Beverly Hunter, and Emma Karel is appreciated. This work was supported by National Institutes of Health, National Institute of Allergy and Infectious Disease Grant R01 AI053692. K. Morse was supported in part by a National Institutes of Health, National Institute of General Medical Sciences pre-doctoral fellowship in Protein Biotechnology (T32 GM008336).
References
- Abbott JR, Palmer GH, Kegerreis KA, Hetrick PF, Howard CJ, Hope JC, Brown WC. Rapid and long-term disappearance of CD4+ T lymphocyte responses specific for Anaplasma marginale major surface protein-2 (MSP2) in MSP2 vaccinates following challenge with live A. marginale. J Immunol. 2005;174:6702–6715. doi: 10.4049/jimmunol.174.11.6702. [DOI] [PubMed] [Google Scholar]
- Alizadeh Z, Karrow N, Mallard BA. Biological effect of varying peptide binding affinity to the BoLA-DRB3*2703 allele. Genet Sel Evol. 2003;35(Suppl 1):S51–65. doi: 10.1186/1297-9686-35-S1-S51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amills M, Ramiya V, Norimine J, Lewin HA. The major histocompatibility complex of ruminants. Re Sci Tech. 1998;17:108–120. doi: 10.20506/rst.17.1.1092. [DOI] [PubMed] [Google Scholar]
- Backert S, Meyer TF. Type IV secretion systems and their effectors in bacterial pathogenesis. Curr Opin Microbiol. 2006;9:207–217. doi: 10.1016/j.mib.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Ballingall KT, Luyai A, Rowlands GJ, Sales J, Musoke AJ, Morzaria SP, McKeever DJ. Bovine leukocyte antigen major histocompatibility complex class II DRB3*2703 and DRB3*1501 alleles are associated with variation in levels of protection against Theileria parva challenge following immunization with the sporozoite p67 antigen. Infect Immun. 2004;72:2738–2741. doi: 10.1128/IAI.72.5.2738-2741.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown WC, Logan KS, Wagner GG, Tetzlaff CL. Cell-mediated immune responses to Babesia bovis merozoite antigens in cattle following infection with tick-derived or cultured parasites. Infect Immun. 1991;59:2418–2426. doi: 10.1128/iai.59.7.2418-2426.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown WC, McGuire TC, Mwangi W, Kegerreis KA, Macmillan H, Lewin HA, Palmer GH. Major histocompatibility complex class II DR-restricted memory CD4+ T lymphocytes recognize conserved immunodominant epitopes of Anaplasma marginale major surface protein 1a. Infect Immun. 2002;70:5521–5532. doi: 10.1128/IAI.70.10.5521-5532.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown WC, Shkap V, Zhu D, McGuire TC, Tuo W, McElwain TF, Palmer GH. CD4+ T-lymphocyte and immunoglobulin G2 responses in calves immunized with Anaplasma marginale outer membranes and protected against homologous challenge. Infect Immun. 1998;66:5406–5413. doi: 10.1128/iai.66.11.5406-5413.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis SA, Ballingall KT. Cattle MHC: evolution in action? Immunol Rev. 1999;167:159–168. doi: 10.1111/j.1600-065x.1999.tb01389.x. [DOI] [PubMed] [Google Scholar]
- Germain RN. Immunology. The ins and outs of antigen processing and presentation. Nature. 1986;322:687–689. doi: 10.1038/322687a0. [DOI] [PubMed] [Google Scholar]
- Germain RN, Quill H. Unexpected expression of a unique mixed-isotype class II MHC molecule by transfected L-cells. Nature. 1986;320:72–75. doi: 10.1038/320072a0. [DOI] [PubMed] [Google Scholar]
- Glass EJ, Oliver RA, Russell GC. Duplicated DQ haplotypes increase the complexity of restriction element usage in cattle. J Immunol. 2000;165:134–138. doi: 10.4049/jimmunol.165.1.134. [DOI] [PubMed] [Google Scholar]
- Haghparast A, Wauben MH, Grosfeld-Stulemeyer MC, van Kooten P, Hensen EJ. Selection of T-cell epitopes from foot-and-mouth disease virus reflects the binding affinity to different cattle MHC class II molecules. Immunogenetics. 2000;51:733–742. doi: 10.1007/s002510000205. [DOI] [PubMed] [Google Scholar]
- Jones GJ, Bagaini F, Hewinson RG, Vordermeier HM. The use of binding-prediction models to identify M. bovis-specific antigenic peptides for screening assays in bovine tuberculosis. Vet Immunol Immunopathol. 2011;141:239–245. doi: 10.1016/j.vetimm.2011.03.006. [DOI] [PubMed] [Google Scholar]
- Lafuente EM, Reche PA. Prediction of MHC-peptide binding: a systematic and comprehensive overview. Curr Pharm Des. 2009;15:3209–3220. doi: 10.2174/138161209789105162. [DOI] [PubMed] [Google Scholar]
- Lewin HA, Russell GC, Glass EJ. Comparative organization and function of the major histocompatibility complex of domesticated cattle. Immunol Rev. 1999;167:145–158. doi: 10.1111/j.1600-065x.1999.tb01388.x. [DOI] [PubMed] [Google Scholar]
- Lopez JE, Palmer GH, Brayton KA, Dark MJ, Leach SE, Brown WC. Immunogenicity of Anaplasma marginale type IV secretion system proteins in a protective outer membrane vaccine. Infect Immun. 2007;75:2333–2342. doi: 10.1128/IAI.00061-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotteau V, Teyton L, Burroughs D, Charron D. A novel HLA class II molecule (DR-sDQ) created by mismatched isotype pairing. Nature. 1987;329:339–341. doi: 10.1038/329339a0. [DOI] [PubMed] [Google Scholar]
- Macmillan H, Norimine J, Brayton KA, Palmer GH, Brown WC. Physical linkage of naturally complexed bacterial outer membrane proteins enhances immunogenicity. Infect Immun. 2008;76:1223–1229. doi: 10.1128/IAI.01356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyasaka T, Takeshima S, Matsumoto Y, Kobayashi N, Matsuhashi T, Miyazaki Y, Tanabe Y, Ishibashi K, Sentsui H, Aida Y. The diversity of bovine MHC class II DRB3 and DQA1 alleles in different herds of Japanese Black and Holstein cattle in Japan. Gene. 2011;472:42–49. doi: 10.1016/j.gene.2010.10.007. [DOI] [PubMed] [Google Scholar]
- Moreno J, Adorini L, Hammerling GJ. Co-dominant restriction by a mixed-haplotype I-A molecule (alpha k beta b) for the lysozyme peptide 52-61 in H-2k x H-2b F1 mice. J Immunol. 1990;144:3296–3304. [PubMed] [Google Scholar]
- Morse K, Norimine J, Palmer GH, Sutten EL, Baszler TV, Brown WC. Association and evidence for linked recognition of type IV secretion system proteins VirB9-1, VirB9-2, and VirB10 in Anaplasma marginale. Infect Immun. 2012;80:215–227. doi: 10.1128/IAI.05798-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassiry MR, Sadeghi B, Tohidi R, Afshari JT, Khosravi M. Comparison of bovine lymphocyte antigen DRB3.2 allele frequencies between two subpopulations of Iranian Holstein cattle. African J Biotechnol. 2008;7:2671–2675. [Google Scholar]
- Noh SM, Brown WC. Adaptive immune responses to infection and opportunities for vaccine development (Anaplasmataceae) In: Palmer GH, Azad AF, editors. Intracellular Pathogens: Rickettsiales. ASM Press; Washington D.C.: 2012. in press. [Google Scholar]
- Norimine J, Brown WC. Intrahaplotype and interhaplotype pairing of bovine leukocyte antigen DQA and DQB molecules generate functional DQ molecules important for priming CD4+ T-lymphocyte responses. Immunogenetics. 2005;57:750–762. doi: 10.1007/s00251-005-0045-6. [DOI] [PubMed] [Google Scholar]
- Norimine J, Han S, Brown WC. Quantitation of Anaplasma marginale major surface protein (MSP)1a and MSP2 epitope-specific CD4+ T lymphocytes using bovine DRB3*1101 and DRB3*1201 tetramers. Immunogenetics. 2006;58:726–739. doi: 10.1007/s00251-006-0140-3. [DOI] [PubMed] [Google Scholar]
- Norimine J, Suarez CE, McElwain TF, Florin-Christensen M, Brown WC. Immunodominant epitopes in Babesia bovis rhoptry-associated protein 1 that elicit memory CD4+-T-lymphocyte responses in B. bovis-immune individuals are located in the amino-terminal domain. Infect Immun. 2002;70:2039–2048. doi: 10.1128/IAI.70.4.2039-2048.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer GH, Brown WC, Rurangirwa FR. Antigenic variation in the persistence and transmission of the ehrlichia Anaplasma marginale. Microbes Infect. 2000;2:167–176. doi: 10.1016/s1286-4579(00)00271-9. [DOI] [PubMed] [Google Scholar]
- Park YH, Joo YS, Park JY, Moon JS, Kim SH, Kwon NH, Ahn JS, Davis WC, Davies CJ. Characterization of lymphocyte subpopulations and major histocompatibility complex haplotypes of mastitis-resistant and susceptible cows. J Vet Sci. 2004;5:29–39. [PubMed] [Google Scholar]
- Pashmi M, Qanbari S, Ghorashi SA, Sharifi AR, Simianer H. Analysis of relationship between bovine lymphocyte antigen DRB3.2 alleles, somatic cell count and milk traits in Iranian Holstein population. J Anim Breed Genet. 2009;126:296–303. doi: 10.1111/j.1439-0388.2008.00783.x. [DOI] [PubMed] [Google Scholar]
- Rupp R, Hernandez A, Mallard BA. Association of bovine leukocyte antigen (BoLA) DRB3.2 with immune response, mastitis, and production and type traits in Canadian Holsteins. J Dairy Sci. 2007;90:1029–1038. doi: 10.3168/jds.S0022-0302(07)71589-8. [DOI] [PubMed] [Google Scholar]
- Sant AJ, Braunstein NS, Germain RN. Predominant role of amino-terminal sequences in dictating efficiency of class II major histocompatibility complex alpha beta dimer expression. Proc Natl Acad Sci USA. 1987;84:8065–8069. doi: 10.1073/pnas.84.22.8065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab AE, Geary TG, Baillargeon P, Schwab AJ, Fecteau G. Association of BoLA DRB3 and DQA1 alleles with susceptibility to Neospora caninum and reproductive outcome in Quebec Holstein cattle. Vet Parasitol. 2009;165:136–40. doi: 10.1016/j.vetpar.2009.07.004. [DOI] [PubMed] [Google Scholar]
- Sharif S, Mallard BA, Wilkie BN, Sargeant JM, Scott HM, Dekkers JC, Leslie KE. Associations of the bovine major histocompatibility complex DRB3 (BoLA-DRB3) alleles with occurrence of disease and milk somatic cell score in Canadian dairy cattle. Anim Genet. 1998;29:185–193. doi: 10.1046/j.1365-2052.1998.00318.x. [DOI] [PubMed] [Google Scholar]
- Silk JD, Schoendorf D, Bartok I, Chai JG, Gray D, Simpson E, Dyson J. Mixed-haplotype MHC class II molecules select functional CD4+ T cells. Mol Immunol. 2005;42:1129–1139. doi: 10.1016/j.molimm.2004.11.024. [DOI] [PubMed] [Google Scholar]
- Spencer JS, Freed JH, Kubo RT. Expression and function of mixed isotype MHC class II molecules in normal mice. J Immunol. 1993;151:6822–6832. [PubMed] [Google Scholar]
- Sutten EL, Norimine J, Beare PA, Heinzen RA, Lopez JE, Morse K, Brayton KA, Gillespie JJ, Brown WC. Anaplasma marginale type IV secretion system proteins VirB2, VirB7, VirB11 and VirD4 are immunogenic components of a protective bacterial membrane vaccine. Infect Immun. 2010;78:1314–1325. doi: 10.1128/IAI.01207-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshima S, Saitou N, Morita M, Inoko H, Aida Y. The diversity of bovine MHC class II DRB3 genes in Japanese Black, Japanese Shorthorn, Jersey and Holstein cattle in Japan. Gene. 2003;316:111–118. doi: 10.1016/s0378-1119(03)00744-3. [DOI] [PubMed] [Google Scholar]
- Tebele N, McGuire TC, Palmer GH. Induction of protective immunity using Anaplasma marginale initial body membranes. Infect Immun. 1991;59:3199–3204. doi: 10.1128/iai.59.9.3199-3204.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unanue ER, Harding CV, Luescher IF, Roof RW. Antigen-binding function of class II MHC molecules. Cold Spring Harb Symp Quant Biol. 1989;54(Pt 1):383–92. doi: 10.1101/sqb.1989.054.01.047. [DOI] [PubMed] [Google Scholar]
- van Eijk MJ, Stewart-Haynes JA, Lewin HA. Extensive polymorphism of the BoLA-DRB3 gene distinguished by PCR-RFLP. Anim Genet. 1992;23:483–496. doi: 10.1111/j.1365-2052.1992.tb00168.x. [DOI] [PubMed] [Google Scholar]
- Xu A, van Eijk MJ, Park C, Lewin HA. Polymorphism in BoLA-DRB3 exon 2 correlates with resistance to persistent lymphocytosis caused by bovine leukemia virus. J Immunol. 1993;151:6977–6985. [PubMed] [Google Scholar]
- Yewdell JW, Bennink JR. The binary logic of antigen processing and presentation to T cells. Cell. 1990;62:203–206. doi: 10.1016/0092-8674(90)90356-j. [DOI] [PubMed] [Google Scholar]
- Zanotti M, Poli G, Ponti W, Polli M, Rocchi M, Bolzani E, Longeri M, Russo S, Lewin HA, van Eijk MJ. Association of BoLA class II haplotypes with subclinical progression of bovine leukaemia virus infection in Holstein-Friesian cattle. Anim Genet. 1996;27:337–341. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.