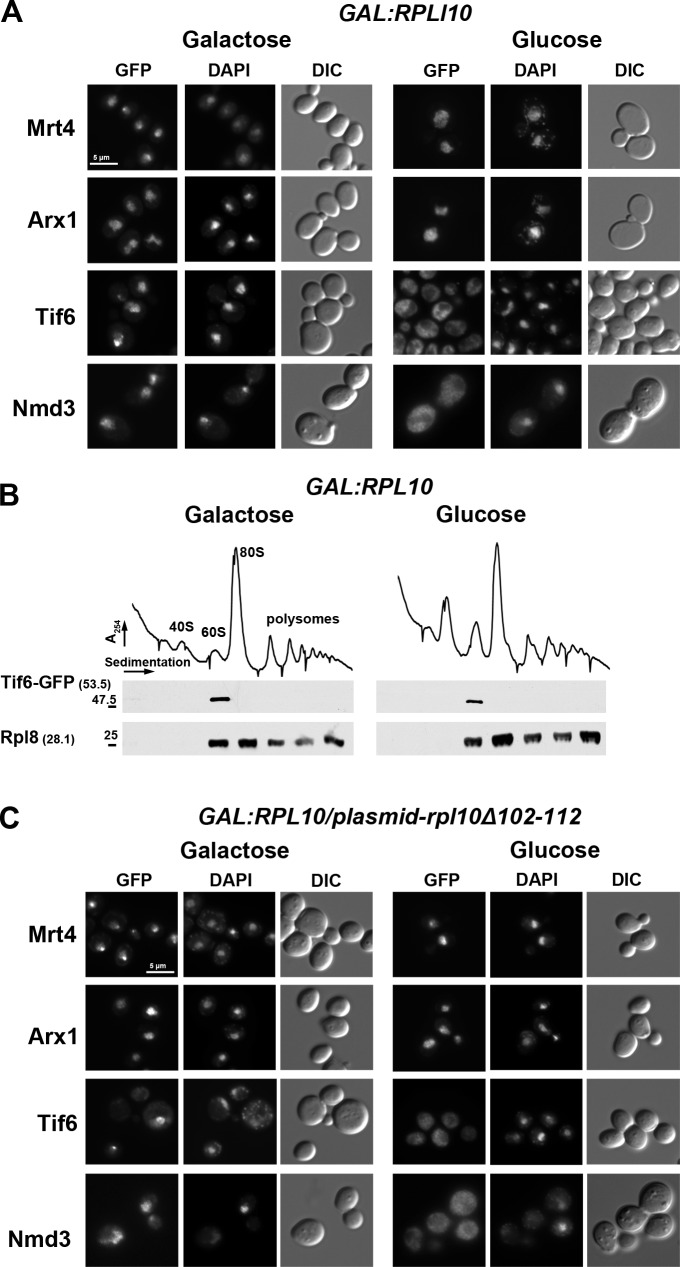
Figure 1.
The P-site loop of Rpl10 is required for the release of Tif6 from 60S ribosomal subunits. (A) The localization of Mrt4, Arx1, Tif6, and Nmd3 was examined in the presence (galactose) or absence (glucose) of ongoing Rpl10 expression. AJY2766 (PGAL1-RPL10 TIF6-GFP), AJY2767 (PGAL1-RPL10 ARX1-GFP), and AJY2768 (PGAL1-RPL10 MRT4-GFP) were grown in galactose to mid-log phase; the cultures were split in two, and for one, Rpl10 expression was repressed for 2 h by the addition of glucose. GFP-tagged proteins were visualized by microscopy. AJY1837 (PGAL1-RPL10 NMD3-GFP crm1-T539C) was treated, as previously described, with the addition of LMB after glucose addition. DIC, differential interference contrast. (B) Sucrose gradient sedimentation of Tif6. AJY2766 (PGAL1-RPL10 TIF6-GFP) was cultured as described in A. Crude extracts were prepared and fractionated by sucrose gradient sedimentation. The position of Tif6 in gradients was monitored by Western blotting using anti-GFP antibody. Anti-Rpl8 was used to monitor the position of 60S subunits. Numbers in parentheses indicate the predicted molecular mass in kilodaltons of the protein being detected. Sizes (kD) and positions (−) of nearest molecular mass markers are indicated. (C) The Rpl10 P-site loop is required for release of Tif6 from 60S subunits. The GFP-tagged strains described in A were transformed with a vector (pAJ1777) expressing mutant RPL10 deleted of the P-site loop (rpl10-Δ102-112). GFP fluorescence of the tagged proteins was monitored under conditions of Rpl10 expression (galactose) or repression (glucose), as in A. Bars, 5 µm.