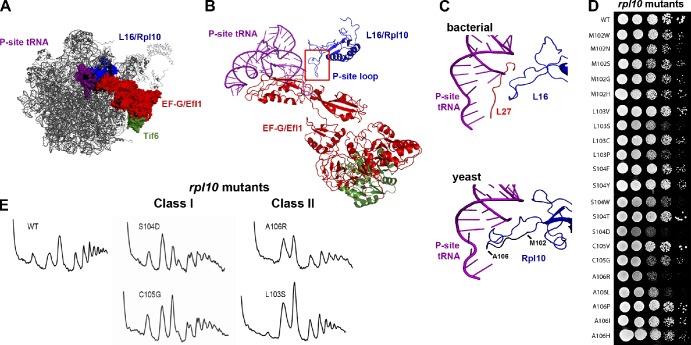
Figure 2.
Mutagenesis of the P-site loop of Rpl10. (A) A composite image of the LSU showing the expected relative positions of L16/Rpl10, P-site tRNA, EF-G/Efl1, and Tif6 was made by docking yeast Tif6 (Protein Data Bank accession no. 2X7N; Gartmann et al., 2010) onto the bacterial 50S subunit with EF-G (PDB accession nos. 2WRI/2WRJ; Gao et al., 2009). The similarities between L16 and Rpl10 and EF-G and Efl1 suggest that we can use the bacterial proteins as proxies for the eukaryotic structures. (B) Ribbon diagram of the molecular linkage between the P-site loop of Rpl10 and Tif6, derived from A. (C) Comparison of the P-site tRNA interactions of L16 and Rpl10. (top) Bacterial L16, L27, and P-site tRNA (adapted from PDB accession nos. 2WRI/2WRJ; Gao et al., 2009). (bottom) Yeast Rpl10 (blue) and P-site tRNA (purple; PDB accession nos. 3IZC/3IZB/3IZE/3IZF; Armache et al., 2010). Residues of the P-site loop that were targeted for mutation (M102 through A106) are shown in black. Isolated mutations are listed. (D) Growth assay of the rpl10 P-site loop mutants. 10-fold serial dilutions of AJY1437 (rpl10Δ::KanMX) containing either WT (pAJ2522) or P-site loop mutants as the sole source of Rpl10 were spotted onto yeast extract peptone dextrose and grown for 2 d at 30°C. (E) The rpl10 P-site loop mutants can be separated in two classes based on polysome profiles. Extracts were prepared from AJY1437 containing WT (pAJ2522) or P-site loop mutants and sedimented through 7–47% sucrose gradients.