Table 2. SBML model definition files that have been used in this article.
| F1 | enzymekinetics1.xml | Enzyme with Michaelis-Menten kinetics, 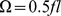 . . |
| F2 | enzymekinetics2.xml | Enzyme with Michaelis-Menten kinetics, 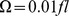
|
| F3 | coopkinetics1.xml | Multi-subunit enzyme with cooperative kinetics, 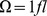
|
| F4 | coopkinetics2.xml | Multi-subunit enzyme with cooperative kinetics, 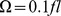
|
| F5 | coremodel1.xml | Circadian clock model, 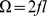 , weak negative feedback , weak negative feedback |
| F6 | coremodel2.xml | Circadian clock model, 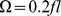 , weak negative feedback , weak negative feedback |
| F7 | coremodel3.xml | Circadian clock model, 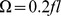 , strong negative feedback , strong negative feedback |
All files and the software iNA are available from the URL http://code.google.com/p/intrinsic-noise-analyzer.
