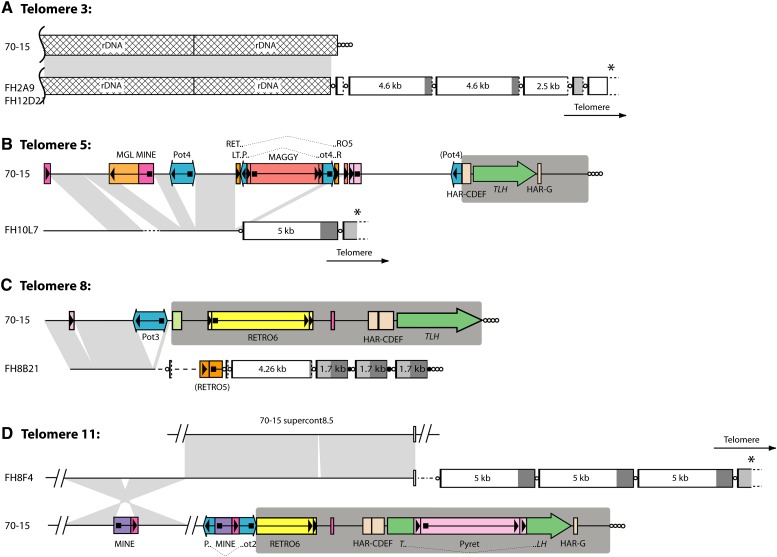
Figure 3 .
Comparison of homologous chromosome ends in strains 70-15 and FH. Panels A\x{2013}D show alignments of four pairs of chromosome ends. Telomere and telomere-like sequences are represented with circles, and the telomere-embedded repeats are depicted as boxes as described in the legend to Figure 2. The TLH regions in the 70-15 subtelomeres are highlighted with dark gray background shading. For cosmid clones that did not contain the terminal telomere repeat array, the direction of the telomere is indicated. Transposon insertions and other repeats in the subtelomere regions are represented by colored boxes with arrows or arrowheads indicating their orientations. Features that are described in the text are labeled. Regions of alignment between the 70-15 chromosome and the FH homolog are connected by light gray shading. Sequences linked to telomere 11 of FH align with two sequences that are well separated in the 70-15 genome, and so two alignments are shown. Sequences that were present near the FH telomeres but absent from the 70-15 genome are drawn as dotted lines, with different line styles representing different sequences. The star-shaped area of shading between telomere 11 homologs represents an inversion that contains a transposon insertion, which in turn caused a deletion in the recipient chromosome. All features are drawn to scale with the exception of the circles that represent the telomeres and telomere-like sequences.