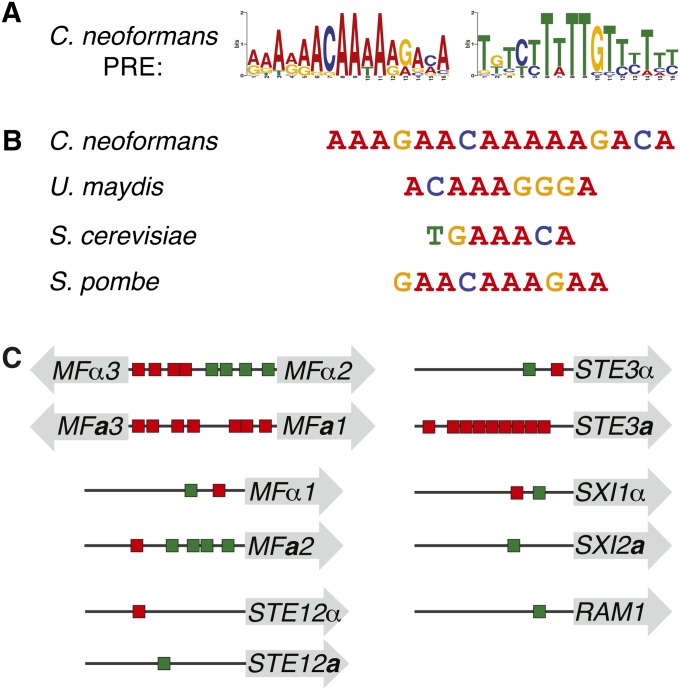
Figure 3 .
The C. neoformans PRE sequence is similar to those of other fungi. (A) Logo of the C. neoformans PRE generated by MEME, shown in forward and reverse complement permutations, with heights of letters corresponding to incidence of a given base at that position (WebLogo assembled from PREs listed in Table 1). The PRE shows no strand bias upstream of target genes and appears in both the A-rich and T-rich permutations on the 5′ strand upstream of target genes. (B) The C. neoformans consensus PRE and those of previously described fungi (Dolan et al. 1989; Sugimoto et al. 1991; Urban et al. 1996; Sahni et al. 2009). (C) Schematic of the PRE content upstream of target genes of particular interest. Shaded arrows represent open reading frames, horizontal lines represent intergenic spaces, and red and green rectangles are representative of PREs, with colors corresponding to direction of PRE as pictured in A. Mating pheromone and receptor genes harbored the most PREs in their upstream regions. Interestingly, orthologous mating-type-specific genes contain differential PRE content, indicating potential regulatory divergence between mating types (e.g., STE3a vs. STE3α).