Abstract
Although neurons are highly polarized, how neuronal polarity is generated remains poorly understood. An evolutionarily conserved inositol-producing enzyme myo-inositol monophosphatase (IMPase) is essential for polarized localization of synaptic molecules in Caenorhabditis elegans and can be inhibited by lithium, a drug for bipolar disorder. The synaptic defect of IMPase mutants causes defects in sensory behaviors including thermotaxis. Here we show that the abnormalities of IMPase mutants can be suppressed by mutations in two enzymes, phospholipase Cβ or synaptojanin, which presumably reduce the level of membrane phosphatidylinositol 4,5-bisphosphate (PIP2). We also found that mutations in phospholipase Cβ conferred resistance to lithium treatment. Our results suggest that reduction of PIP2 on plasma membrane is a major cause of abnormal synaptic polarity in IMPase mutants and provide the first in vivo evidence that lithium impairs neuronal PIP2 synthesis through inhibition of IMPase. We propose that the PIP2 signaling regulated by IMPase plays a novel and fundamental role in the synaptic polarity.
Keywords: bipolar disorder, Caenorhabditis elegans, myo-inositol monophosphatase (IMPase), synapse, lithium
NEURONS ensure polarized information flows with their diverse morphology. A typical mammalian neuron receives synaptic inputs at branching dendrites and sends signals through a long projecting axon. It has been shown that phosphoinositides, which are derived from combinational phosphorylation of phosphatidylinositol (PI), play important roles in neuronal polarity (Arimura and Kaibuchi 2005; Arimura and Kaibuchi 2007; Skwarek and Boulianne 2009). PI is synthesized from myo-inositol, which is supplied by uptake from the extracellular environment, de novo synthesis from glucose, or recycling from phosphoinositides (Figure 3B; Agam et al. 2009). Of these, the de novo synthesis and recycling pathways require myo-inositol monophosphatase (IMPase), an evolutionarily conserved enzyme that produces inositol by dephosphorylating inositol monophosphate (Figure 3B; Agam et al. 2009).
Figure 3.
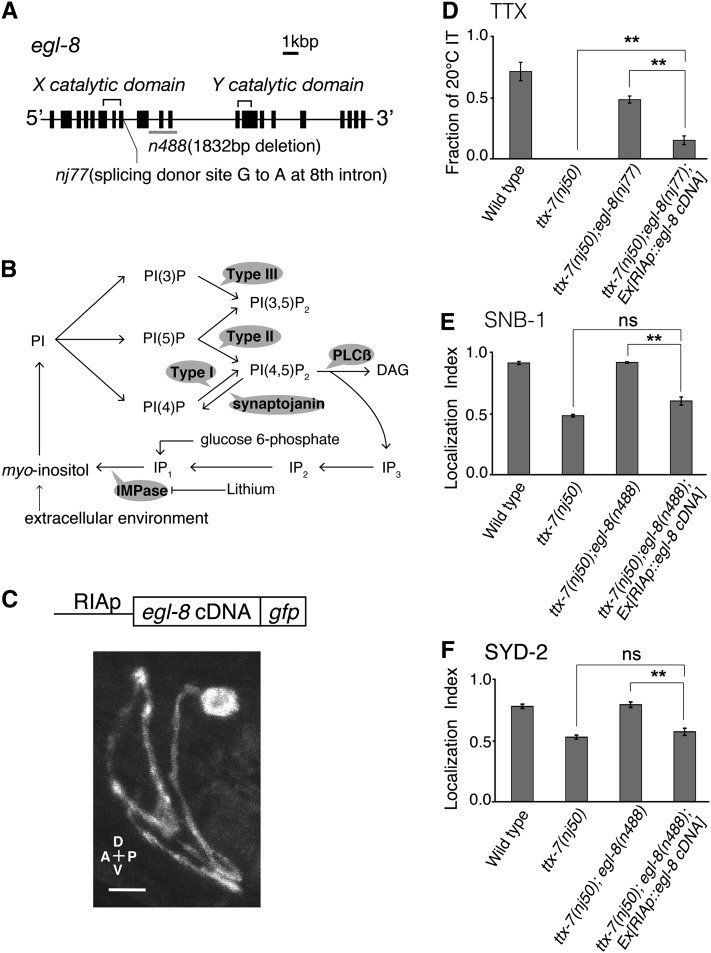
Loss of EGL-8 function in RIA suppresses the abnormalities in ttx-7 mutants. (A) Locations of nj77 and n488 mutations in egl-8 gene. Solid boxes indicate exons. (B) Simplified model of PI metabolism. myo-Inositol is supplied by de novo synthesis from glucose 6-phosphate and recycling from phosphoinositides, which require IMPase to dephosphorylate inositol monophosphate (IP1). myo-Inositol is also supplied from the extracellular environment via membrane-associated transporters. PI is synthesized from myo-inositol, and type I, II, and III of PIP kinases phosphorylate PI(4)P, PI(5)P, and PI(3)P, respectively. PLCβ cleaves PI(4,5)P2 into DAG and IP3. IP3 is sequentially dephosphorylated into myo-inositol. Synaptojanin dephosphorylates the five-position phosphate from PI(4,5)P2. IMPase can be inhibited by lithium in vivo. (C) The subcellular localization of EGL-8::GFP in the RIA neuron. EGL-8::GFP was not localized to a specific region. Scale bar, 5 µm. (D) Expressing egl-8 cDNA specifically in RIA reduced the fraction of IT behavior in ttx-7(nj50);egl-8(nj77) double mutants. About 20 animals cultivated at 20° were examined in more than three trials, which were compared in ANOVA. (E and F) The localization indices of SNB-1::VENUS (E) and GFP::SYD-2 (F). The RIA-specific expression of egl-8 cDNA reduced the localization indices in ttx-7(nj50);egl-8(n488) double mutants. Steel–Dwass multiple comparison tests were performed (n ≥ 11 animals).
The in vivo function of IMPase has been extensively explored because of its sensitivity to lithium, a drug for bipolar disorder (Cade 1949). Despite the clinical usage for more than half of a century, how lithium exerts its therapeutic effect in patient brains is still enigmatic. Currently, a dominant explanation for the action of lithium is exemplified by the “inositol depletion hypothesis.” This hypothesis, mainly based on in vitro studies, holds that IMPase inhibition by lithium limits production of inositol, thereby dampening phosphatidylinositol 4,5-bisphosphate (PIP2)-mediated signaling (Supporting Information, Figure S2; Berridge et al. 1982; Berridge et al. 1989; Schloesser et al. 2008; Machado-Vieira et al. 2009). Although it has been shown that lithium inhibits IMPase (Hallcher and Sherman 1980; Hedgepeth et al. 1997) and reduces inositol levels (Maslanski et al. 1992; O’donnell et al. 2000), whether the reduction of inositol affects neuronal PIP2 levels in vivo has been disputed (Batty and Downes 1994; Dixon et al. 1994; Berry et al. 2004; Schloesser et al. 2008; King et al. 2009; Machado-Vieira et al. 2009). The oppositions to the hypothesis are mainly based on two reasons. First, since inositol can be supplied from the extracellular environment independently of IMPase, inhibition of IMPase by lithium might cause only a marginal reduction in the inositol level, which might not substantially impair the synthesis of PI (Batty and Downes 1994; Berry et al. 2004). Second, since inhibition of IMPase might alter the levels of inositol polyphosphates that are important regulators of gene expression in vivo (Odom et al. 2000; Shaldubina et al. 2002; Seeds et al. 2005; Lee et al. 2007), lithium could exert its effect by interfering with these PI-independent metabolic pathways. Thus, it remains unclear how lithium exerts its effect in vivo.
We previously reported that the ttx-7 gene, the Caenorhabditis elegans ortholog of IMPase, is required for sensory behaviors. The behavioral defects of ttx-7 mutants result from abnormality in polarized localization of both pre- and postsynaptic proteins in the interneuron named RIA (Tanizawa et al. 2006). The exogenous application of lithium to wild-type animals elicited both the synaptic and behavioral defects similar to those in ttx-7 mutants (Tanizawa et al. 2006), suggesting that lithium inhibits the C. elegans IMPase. However, it remains unknown how the inhibition of IMPase leads to such defects.
In this study, we conducted a genetic suppressor screen for ttx-7 mutants. We found that mutations in the gene egl-8, which encodes a homolog of phospholipase Cβ (PLCβ) (Lackner et al. 1999; Miller et al. 1999), strongly suppress both the synaptic and behavioral defects of ttx-7 mutants. Since PLCβ cleaves PIP2, this observation suggests that the accumulation of PIP2 corrected the defects. Indeed, through screening for known inositol metabolic genes, we found that a mutation in the unc-26 gene, a homolog of human synaptojanin 1 that dephosphorylates PIP2 (Cremona et al. 1999; Harris et al. 2000), also suppresses the synaptic defect in ttx-7 mutants. Further, egl-8 mutants showed strong resistance to the lithium treatment. Thus, these results provide the first genetic evidence that disruption of IMPase by lithium affects PIP2 levels in neurons of living animals and suggest that the PIP2 signaling establishes polarized localization of pre- and postsynaptic components in vivo.
Materials and Methods
Strains and genetics
C. elegans cultures were maintained essentially as described (Brenner 1974). The following strains were used: wild-type Bristol strain (N2), wild-type Hawaiian strain (CB4856) for mapping with snip-SNPs method (Wicks et al. 2001), CB47 unc-11(e47) I, EG3361 gqIs25[rab-3p::ppk-1, lin-15(+)] I, IK575 ttx-7(nj40) I, IK589 ttx-7(nj50) I, IK685 njIs20[glr-3p::syd-2::gfp, rol-6gf] I, IK765 njIs16[glr-3p::eat-4::gfp, glr-3p::snb-1::dsredmonomer, rol-6gf] I, CB1265 unc-104(e1265) II, CB205 unc-26(e205) IV, IK661 njIs9[glr-3p::snb-1::venus, ofm-1p::gfp] IV, IK718 njIs12[glr-3p::glr-1::gfp, ges-1p::Dsredmonomer] V, IK777 egl-8(nj77) V, MT1083 egl-8(n488) V, and multiple mutants or transgenic strains generated form them. The rest of strains used are listed in Table 1.
Table 1. Localizarion of SNB-1 in mutants related to neuronal polarity, synapse formation, or inositol metabolism.
| Strain used | Mutation | Gene | Localization of snb-1 |
|---|---|---|---|
| N2 | Wild type | — | Presynaptic region |
| IK589 | ttx-7(nj50) | IMPase | Entire process |
| Polarity- or synapse-related genes | |||
| KU17 | lrk-1(km17)a | LRRK2/PARK8-related kinase | WT |
| VC898 | cdc-42(gk388)b,c | Cdc42 | WT |
| NG324 | wsp-1(gm324b | WASP | WT |
| VC2053 | wip-1(ok2435)b,c | WASP-interacting protein | WT |
| VC2706 | wve-1(ok3308)b,c | WAVE | WT |
| DR1 | unc-101(m1) | AP-1 medium subunit | WT |
| DR1 | ttx-7(nj40) unc-101(m1) | AP-1 medium subunit | ttx-7-like |
| CB1193 | unc-33(e1193) | CRMP | WT |
| CB204 | unc-33(e204) | CRMP | WT |
| SP1382 | unc-33(mn407) | CRMP | WT |
| EM67 | mab-20(bx24)a | Semaphorin-2A | WT |
| CB78 | unc-6(e78)a | Netrin | WT |
| CB271 | unc-40(e271)a | Netrin receptor | WT |
| CB362 | unc-44(e362)b | Ankyrin G | WT |
| Inositol metabolism-related genes | |||
| CB205 | unc-26(e205) | Synaptojanin | WT (Figure 4) |
| CB205 | ttx-7(nj50);unc-26(e205) | Synaptojanin | WT (Figure 4) |
| RB1535 | arf-1.1(ok1840) | ADP-ribosylation factor family | WT |
| VC567 | arf-1.2(ok796) | ADP-ribosylation factor family | WT |
| FX1447 | arf-6(tm1447)b | ADP-ribosylation factor family | WT |
| KU22 | pld-1(km22) | Phospholipase D | WT |
| VC1587 | ocrl-1(gk752) | OCRL | WT |
| VC1587 | ttx-7(nj40) ocrl-1(gk752) | OCRL | ttx-7-like |
| IK1130 | age-1(mg305) | Phosphoinositide 3-kinase | WT |
| IK1130 | ttx-7(nj50);age-1 (mg305) | Phosphoinositide 3-kinase | ttx-7-like |
| RB1813 | piki-1(ok2346) | Phosphoinositide 3-kinase | WT |
| KR1440 | vps-34(h797)b,c | Phosphoinositide 3-kinase | WT |
| FX2348 | F35H12.4(tm2348)b | Phosphatidylinositol kinase | WT |
| VC2563 | Y75B8A.24(ok3320)b | Phosphatidylinositol kinase | WT |
| EG3361 | ttx-7(nj50) gpIs25[rab-3p::ppk-1] | Type I PI-4-phosphate kinase | ttx-7-like |
| FX3741 | ppk-2(tm3741)b | Type II PI -5-phosphate kinase | WT |
| MT7531 | ppk-3(n2835)b | Type III PI-3-phosphate kinase | WT |
| MT12352 | trr-1(n3630)b,c | TRAAP subfamily | WT |
| TR1331 | smg-1(r861)b | Phosphatidylinositol kinase | WT |
| VC381 | atm-1(gk186)b | ATM family | WT |
| VC728 | atl-1(ok1063)b | ATM family | WT |
| VC2312 | let-363(ok3013)b,c | FRAP1 | WT |
| FX753 | plc-1(tm753) | Phospholipase c | WT |
| FX753 | ttx-7(nj40);plc-1 (tm753) | Phospholipase c | ttx-7-like |
| RB1496 | plc-2(ok1761)b | Phospholipase c | WT |
| RB1496 | ttx-7(nj50);plc-2(ok1761)b | Phospholipase c | ttx-7-like |
| MT1434 | egl-30(n686) | G-protein α-subunit | WT |
| KY26 | egl-30(tg26gf) | G-protein α-subunit | WT |
| MT1434 | egl-30(n686) ttx-7(nj50) | G-protein α-subunit | ttx-7-like |
Localization of SNB-1 in RIA neuron was examined. About 10 adult animals were examined in more than three trials for each genotype. In the mutants without the superscripts, the localization of SNB-1::VENUS expressed from integrated array was observed. WT and ttx-7-like represents the wild-type and ttx-7-like mutant phenotypes of SNB-1 localization, respectively.
The localization of SNB-1::DsRedmonomer expressed from integrated array was observed.
The localization of SNB-1::VENUS expressed from extrachromosomal array was observed.
These mutants display larval arrest or developmental defects. The localization of SNB-1 in these mutants was examined at larval stages; SNB-1 localized to the distal region of the process in wild-type animals at laval stages as well as at the adult stage.
Behavioral assay
The population thermotaxis assay was performed as previously reported (Ito et al. 2006) except for Figure S3, F–G, in which the assay duration was 120 min. The individual thermotaxis assay was performed as described (Mori and Ohshima 1995). The salt chemotaxis assay was performed as described (Komatsu et al. 1996).
Genetic screens for mutations that suppress thermotaxis defects of ttx-7 mutants
ttx-7(nj40) animals were mutagenized with ethyl methanesulfonate (EMS) as described before (Brenner 1974), and F1 progeny was isolated to 6-cm NGM plates. F2 progeny from five F1 plates was cultured at 23° and was assayed in the population thermotaxis assay for 40 min. egl-8(nj77) was isolated as animals that migrated to 23°.
Mapping of nj77
We outcrossed ttx-7(nj40) to CB4856 to generate the strain carrying ttx-7(nj40) in a Hawaiian background. By utilizing the SNPs (single nucleotide polymorphisms) between this strain and the suppressor, we mapped nj77 to a 1 Mbp region of the left end of chromosome V.
Molecular biology
An egl-8 cDNA (KP316) is a gift from Dr. Stephan Nurrish. The promoter of the glr-3 gene was used as an RIA-specific promoter. To generate glr-3p::egl-8 cDNA (pUBA13), the egl-8 cDNA was amplified by PCR from KP316 plasmid, and the ttx-7a cDNA::egfp of glr-3p::ttx-7a cDNA::egfp (pTAN58) was replaced by the egl-8 cDNA. To generate glr-3p::gfp::egl-8 cDNA (pUBA21), an gfp::egl-8 cDNA was amplified by PCR from acr-2p::gfp::egl-8 cDNA (REW1) plasmid, and the ttx-7a cDNA::egfp of glr-3p::ttx-7a cDNA::egfp (pTAN58) was replaced by the gfp::egl-8 cDNA. To generate glr-3p::unc-101 cDNA::egfp (pUBA35), a unc-101 cDNA was amplified from C. elegans yeast two-hybrid cDNA library (Cosmo Bio Co., Ltd), and the ttx-7a cDNA of glr-3p::ttx-7a cDNA::egfp (pTAN58) was replaced by the unc-101 cDNA
Transgenic animals
Germline transformation was performed by co-injecting experimental DNA (1–100 ng/µl) and an injection marker pKDK66 (ges-1p::NLS::GFP), ofm-1::gfp, pRF4 (rol-6gf), or pTAN124.5 (ges-1p::Dsredmonomer) (Mello et al. 1991). Multiple independent transgenic lines were established for each experimental DNA. For comparison of phenotypes on different genetic backgrounds, transgenic arrays were transferred by intercrossing. Strains with integrated arrays were established by TMP/UV mutagenesis of animals carrying an extrachromosomal array as described (Tanizawa et al. 2006).
Lithium treatment
Animals were cultivated on LiCl-containing NGM plates from birth. LiCl (Wako) was added at 15 mM concentration to NGM medium. We used 1-day-old adults for phenotypic analyses.
Observation and quantification of synaptic molecule localization
An Axioplan2 light microscope (Zeiss) was used to observe the synaptic molecule localization. The fluorescent images were captured with a confocal laser-scanning microscope FV1000 (Olympus). The quantifications of localization indices for SNB-1::VENUS, GFP::SYD-2, and GLR-1::GFP were performed on adult animals with integrated arrays. The localization index was calculated using ImageJ software (NIH): the area and mean fluorescence intensity of the background, presynaptic region (region A), and non-presynaptic region (region B) of the RIA process (Figure 2B) were measured for each slice of a confocal image. The total intensity in each region for each slice was generated by subtracting the mean intensity of the background from that of the region of interest, which was then multiplied by its area. The fluorescence of the region A and B was calculated by summing the total intensity of each slice of each region. The localization index was calculated as fluorescence A / fluorescence A + B.
Figure 2.
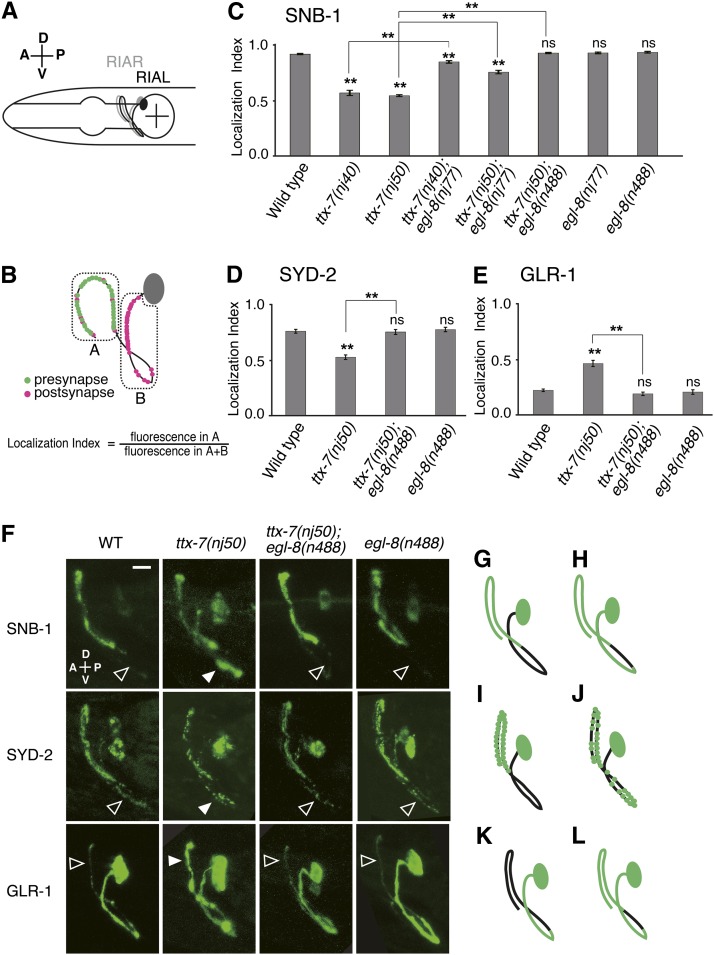
Mutations in egl-8 strongly suppress the synaptic defects of ttx-7 mutants. (A) Schematic of the head region of C. elegans and a pair of RIA. (B) Schematic of pre- and postsynapses distribution in RIA (White et al. 1986). The localization index was calculated using the equation shown here. Measurement of fluorescent intensity was performed as described in Materials and Methods. (C–E) Comparison of the localization indices of SNB-1::VENUS (C), GFP::SYD-2 (D), and GLR-1::GFP (E) in the RIA neuron in each genotype. Mutations in egl-8 strongly suppressed the localization defects of the synaptic proteins in ttx-7 mutants. Note that egl-8(n488) suppressed more strongly than egl-8(nj77) (C). Steel–Dwass multiple comparison tests were performed (n ≥ 11 animals). The marks on the bars of each genotype indicate comparisons with wild type. The marks on the lines indicate comparisons between indicated genotypes. (F) Representative images of the distribution of each synaptic protein in the RIA neuron in each genotype. Solid arrowheads indicate ectopic fluorescence, and open arrowheads indicate absence of the ectopic fluorescence. Scale bar, 5 µm. (G and H) Schematic of SNB-1::VENUS localization in wild-type animals, ttx-7(nj50);egl-8(n488) and egl-8(n488) mutants (G) and that in ttx-7(nj50) mutants (H). SNB-1::VENUS is mislocalized at the proximal region of the process in ttx-7(nj50) mutants. (I and J) Schematic of GFP::SYD-2 localization in wild-type animals, ttx-7(nj50);egl-8(n488) and egl-8(n488) mutants (I) and that in ttx-7(nj50) mutants (J). GFP::SYD-2 mainly localized to the distal region of RIA in wild-type animals, ttx-7(nj50);egl-8(n488) and egl-8(n488) mutants, while it dispersed in whole process in ttx-7(nj50) mutants. (K and L) Schematic of GLR-1::GFP localization in wild-type animals and ttx-7(nj50);egl-8(n488) and egl-8(n488) mutants (K) and that in ttx-7(nj50) mutants (L). GLR-1::GFP mainly localized in the proximal region of RIA in wild-type animals, ttx-7(nj50);egl-8(n488) and egl-8(n488) mutants, while it diffused in whole process in ttx-7(nj50) mutants.
Statistics
Error bars in all figures indicate standard error of the mean (SEM). We treated thermotaxis indices and localization indices as parametric and nonparametric data, respectively. The comparison test methods applied are indicated in each figure legend. The double asterisks (**), single asterisks (*), and no significances (NS) in all figures represent P < 0.01, P < 0.05, and P > 0.05, respectively.
Results
A mutation in the egl-8 gene strongly suppresses the behavioral abnormalities of ttx-7 mutants
The ttx-7 gene encodes the sole ortholog of IMPase gene in C. elegans. Tanizawa et al. (2006) showed that the loss of TTX-7 causes defects in both polarized distribution of synaptic proteins in the RIA interneuron and behaviors including thermotaxis (Tanizawa et al. 2006). Thermotaxis is one of the most characterized experience-dependent behaviors in C. elegans: when well-fed animals cultivated at a certain temperature are placed on a temperature gradient (shallower than 1°/cm) without food, they migrate toward their cultivated temperatures and move isothermally (Hedgecock and Russell 1975; Mori and Ohshima 1995; Mohri et al. 2005; Jurado et al. 2010). RIA receives synaptic inputs from upstream interneurons in the neural circuit regulating thermotaxis (Figure S1A; Mori and Ohshima 1995). Since RIA neuron-specific expression of ttx-7 cDNA rescued both the synaptic and thermotaxis defects, abnormal thermotaxis phenotype of ttx-7 mutants is likely caused by synaptic defects in RIA (Tanizawa et al. 2006).
To clarify further how IMPase regulates the synaptic polarity and consequently sensory behaviors, we conducted a genetic suppressor screen for ttx-7 mutants utilizing a population thermotaxis assay (Materials and Methods). Of five isolates among ∼2000 genomes screened in a ttx-7(nj40) background, we focused on the mutation nj77 that strongly suppressed the thermotaxis defect.
The snip-SNPs method (Wicks et al. 2001) and subsequent sequencing analyses revealed a G-to-A mutation in the splicing donor site of the eighth intron of the egl-8 gene in the nj77 mutant genome (Figure 3A). The egl-8 gene encodes a homolog of PLCβ, most closely related to PLCβ4 in vertebrates (Lackner et al. 1999; Miller et al. 1999). ttx-7(nj40);nj77 mutants showed slightly flattened sinusoidal tracks on cultivation plates unlike ttx-7(nj40) single mutants, which are characteristic of egl-8 mutant animals (data not shown; Lackner et al. 1999). RIA-specific expression of egl-8 cDNA abolished the suppressible effect of nj77 mutation (Figure 3D and Figure S1D; discussed below). These results indicate that nj77 is an allele of egl-8.
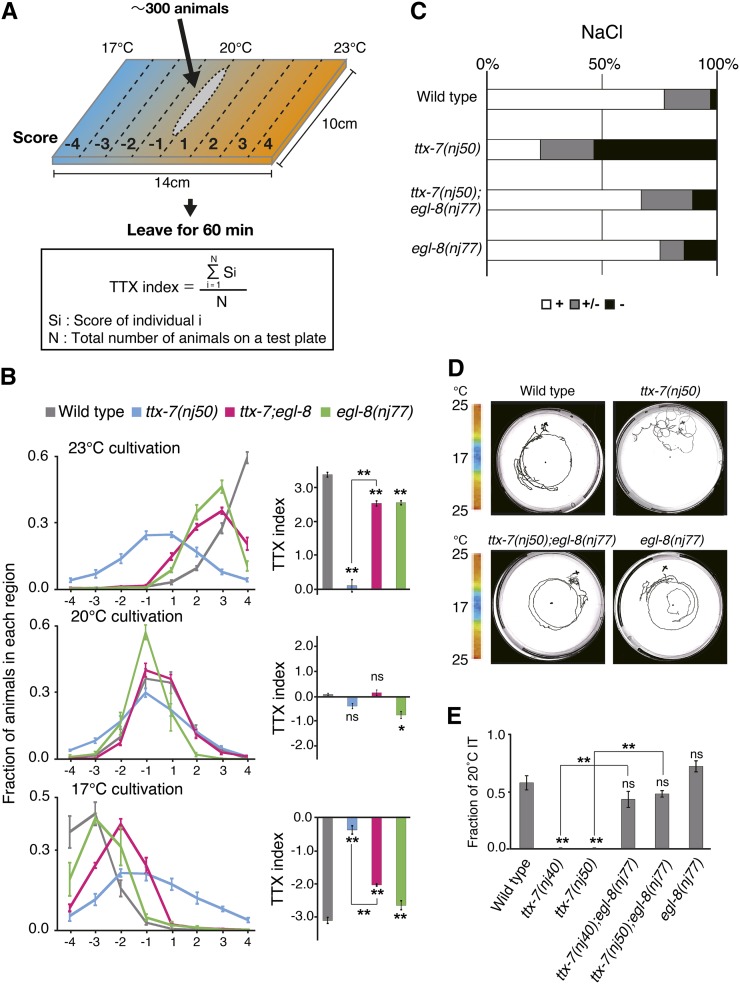
We further analyzed the thermotaxis behavior of ttx-7;egl-8(nj77) mutants, using a deletion or a hypomorphic allele, ttx-7(nj50) or ttx-7(nj40), respectively. To investigate the ability of migrating toward cultivation temperature on a thermal gradient, we utilized the population thermotaxis assay (Figure 1A; Ito et al. 2006). After cultivation at 23°, 20°, or 17° in a well-fed condition, most wild-type animals migrated toward their cultivation temperatures, whereas ttx-7 mutants dispersed on the assay plate (Figure 1B and Figure S1B). In contrast, ttx-7;egl-8(nj77) mutants migrated toward their cultivation temperatures. ttx-7;egl-8(nj77) and egl-8(nj77) single mutants accumulated at the temperature slightly higher or lower than wild-type animals in 17° or 23° cultivation, respectively (Figure 1B and Figure S1B). Since animals carrying a deletion allele of egl-8, egl-8(n488), do not move on the assay plate owing to locomotion defects (Okochi et al. 2005), the thermotaxis abnormalities in ttx-7;egl-8(nj77) and egl-8(nj77) mutants might be caused by a defect in locomotion. To test this possibility, we assayed wild-type and egl-8(nj77) mutant animals cultivated at 17° or 23° on the assay plates without the temperature gradient and compared the TTX deviations, which reflect the dispersion of the animals (Figure S3A; Ito et al. 2006). The TTX deviations between wild-type and egl-8(nj77) mutants were not considerably different (Figure S3, B–E). We found that extending the assay duration from 60 to 120 min improved the thermotaxis performance of egl-8(nj77) mutants cultured at 17° but not at 23° (Figure S3, F and G). These results suggest that egl-8 is involved in thermotaxis rather than merely affecting the locomotion.
Figure 1.
Behavior of ttx-7;egl-8 double mutants. (A) Procedure for the population thermotaxis assay. Between 50 and 300 animals cultivated at a certain temperature were placed at the center of the liner temperature gradient ranging from 17° to 23° in 14 cm width. After 60 min, the number of animals at each region was counted. The TTX index was calculated using the equation shown here. (B) Distributions and TTX indices of animals cultivated at 17°, 20°, and 23°. While ttx-7(nj50) mutants showed almost athermotactic behavior, ttx-7(nj50);egl-8(nj77) double mutants migrated toward the cultivation temperatures. Tukey–Kramer test was applied (n ≥ 4 assays). The marks on the bars of each genotype represent comparisons with wild type. The marks on the lines represent comparisons between indicated genotypes. (C) Individual chemotaxis assay to NaCl. (+) strong attraction; (+/−) modest attraction; (−) no attraction to NaCl. egl-8(nj77) strongly suppressed the chemotaxis defect of ttx-7(nj50) mutants. n ≥ 57 animals. (D) Individual thermotaxis assay of animals cultivated at 20°. The center and edge of the 9-cm plate are maintained at 17° and 25°, respectively. In contrast to the random movement of ttx-7(nj50) mutants, ttx-7(nj50);egl-8(nj77) mutants showed clear isothermal tracking (IT) around 20° as well as wild-type animals. (E) Fraction of animals that moved isothermally around 20° in the individual thermotaxis assay. nj40 and nj50 are hypomorphic and putative null alleles for ttx-7, respectively. egl-8(nj77) strongly suppressed the defect of ttx-7 mutants. About 20 animals were examined in more than three trials, which were compared in ANOVA. The marks on the bars of each genotype indicate comparisons with wild type. The marks on the lines represent comparisons between indicated genotypes.
After reaching the cultivation temperature, animals move isothermally (IT behavior) (Hedgecock and Russell 1975; Mori and Ohshima 1995; Ryu and Samuel 2002; Luo et al. 2006). Since RIA is essential for IT behavior (Figure S1A; Ohnishi et al. 2010), we tested this behavior using the individual thermotaxis assay with a radial temperature gradient (Mori and Ohshima 1995). In contrast to wild-type animals showing clear isotherms, ttx-7 mutants moved almost randomly on the gradient (Figure 1, D and E). By contrast, ttx-7;egl-8(nj77) double mutants showed IT behavior similar to those of wild-type animals and egl-8(nj77) single mutants (Figure 1, D and E), suggesting that egl-8(nj77) restores the function of RIA in ttx-7 mutants.
ttx-7 mutants was previously shown to be defective in salt attraction (Figure 1C; Tanizawa et al. 2006). We found that ttx-7(nj50);egl-8(nj77) and egl-8(nj77) mutants were attracted to NaCl to the similar extent to that of wild-type animals (Figure 1C). Taken together, we concluded that egl-8(nj77) confers strong suppression for the behavioral defects of ttx-7 mutants.
Loss of EGL-8 suppresses the synaptic defects of ttx-7 mutants
We next addressed whether egl-8 mutations also suppress the synaptic defects of ttx-7 mutants. The RIA interneuron has a single process where pre- and postsynaptic regions are segregated, providing a unique platform from which to analyze the polarized distribution of synaptic proteins in vivo (Figure 2, A and B; White et al. 1986; Tanizawa et al. 2006; Margeta et al. 2009). The localization of fluorescent marker-tagged synaptic proteins in the process was evaluated using the “localization index” shown in Figure 2B. The synaptic vesicle-associated protein SNB-1 fused to VENUS was exclusively localized to the presynaptic region of RIA in wild-type and egl-8 mutant animals (Figure 2, C, F, and G). As previously reported, SNB-1::VENUS was abnormally localized in both pre- and nonpresynaptic regions of the RIA process in ttx-7 mutants (Figure 2, C, F, and H; Tanizawa et al. 2006), whereas both egl-8(nj77) and egl-8(n488) markedly suppressed the localization defect of ttx-7 mutants (Figure 2, C, F, and G). We noted that the suppression by egl-8(nj77) was weaker than egl-8(n488) (P < 0.01: comparison between nj50;nj77 and nj50;n488 in Figure 2C). This result and the locomotion phenotype described above suggest that nj77 is a hypomorphic allele of egl-8. We next examined the localization of SYD-2, a presynaptic active zone protein (Yeh et al. 2005). The SYD-2 tagged with GFP was mainly localized to the presynaptic region in wild-type and egl-8 mutant animals, whereas in ttx-7 mutants, the fluorescent puncta in the presynaptic region was dispersed throughout the whole process (Figure 2, D, F, I, and J; Tanizawa et al. 2006). As in the case of SNB-1, egl-8(n488) strongly suppressed this defect (Figure 2, D, F, and I).
We also examined postsynaptic specializations using GLR-1, an AMPA-type glutamate receptor. The GLR-1::GFP was localized to the postsynaptic region in wild-type and egl-8 mutant animals, while it diffused throughout the entire process in ttx-7 mutants (Figure 2, E, F, K and L; Tanizawa et al. 2006). egl-8(n488) completely suppressed this defect (Figure 2, E, F and K). These results indicate that egl-8 mutations confer the strong suppression for the synaptic defects in ttx-7 mutants.
Disruption of EGL-8-mediated PIP2 degradation in RIA suppresses the abnormalities in ttx-7 mutants
To examine where the egl-8 gene acts, we conducted cell-specific rescue experiments. The RIA-specific expression of egl-8 cDNA in ttx-7;egl-8 double mutants substantially reduced the fraction of IT behavior (Figure 3D and Figure S1D), and also disrupted localizations of SNB-1 and SYD-2 similarly to ttx-7 single mutants (Figure 3, E and F, and Figure S4D). These results indicate that the loss of egl-8 function in RIA restores the abnormalities in ttx-7 mutants.
egl-8 encodes PLCβ that hydrolyzes PIP2 on plasma membrane (Figure 3B; Lackner et al. 1999; Miller et al. 1999). Because local PI metabolism on cell membrane is thought to be important for polarity establishment (Skwarek and Boulianne 2009), we assessed whether the PIP2 hydrolysis is restricted to a domain(s) of the RIA process such as pre- and postsynaptic domains by examining the subcellular localization of EGL-8. A functional GFP::EGL-8 (Figure S1C) was diffusely localized presumably on the membrane of the entire process and cell body (Figure 3C). This result suggests that PIP2 hydrolysis is not restricted to any specific regions of the RIA process but does not exclude the possibility that EGL-8 activity is spatially restricted by a regulator protein. Given that EGL-8 is activated by the G-protein α-subunit EGL-30 at neuromuscular junctions (Lackner et al. 1999; Miller et al. 1999), we speculated that EGL-30 also regulates EGL-8 in RIA. However, we did not see the clear suppression of the defective localization of SNB-1::VENUS in ttx-7(nj50);egl-30(n686) double mutants, and gain- or loss-of-function mutations in egl-30 did not cause localization defects (Table 1). It is still possible that a protein different from EGL-30 regulates the activity of EGL-8 in RIA.
Screening for PI metabolic genes regulating the localization of synaptic components
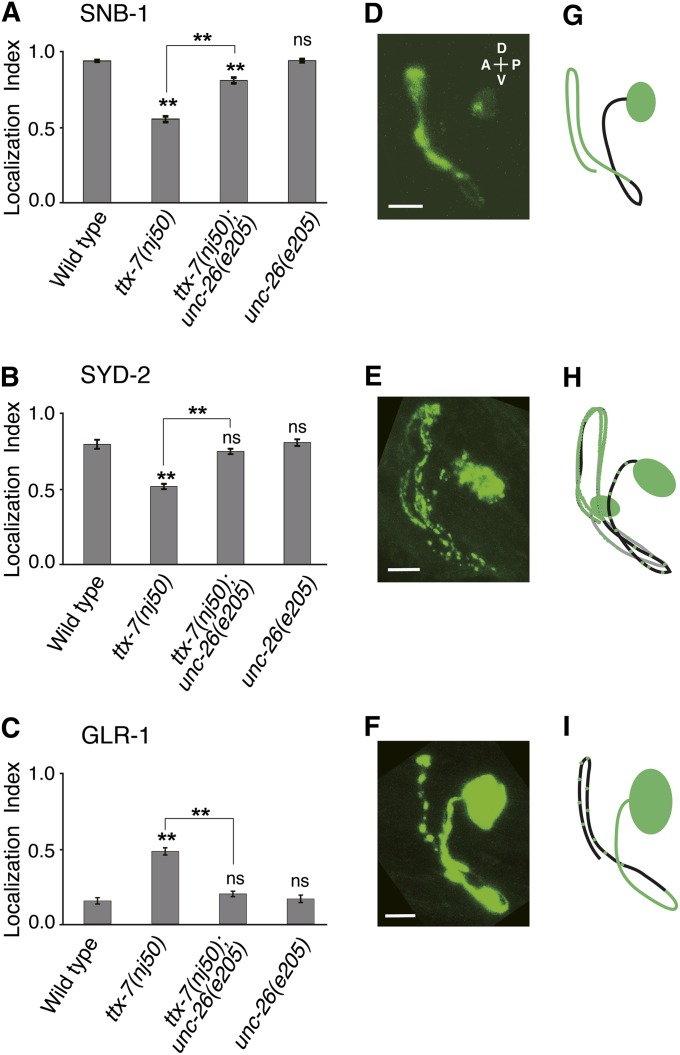
The loss of EGL-8/PLCβ would cause an increase of PIP2 and a decrease of IP3 and DAG (Figure 3B). To test which of these two changes is responsible for the suppression of the ttx-7 phenotype, we examined the SNB-1 localization in the RIA neuron of animals mutant for genes involved in PI metabolism (Table 1). First, we examined the gene unc-26, encoding a C. elegans ortholog of human synaptojanin 1 that regulates the clathrin uncoating step of endocytosis through dephosphorylation of PIP2 on plasma membrane (Figure 3B; Cremona et al. 1999; Harris et al. 2000). The unc-26 mutation substantially suppressed the localization defects of synaptic proteins in ttx-7 mutants (Figure 4 and Table1). We could not assess the thermotaxis phenotype of ttx-7(nj50);unc-26(e205) double mutants owing to locomotion defects. Given that the level of PIP2 was reported to be selectively increased in neurons of synaptojanin-knockout mice (Cremona et al. 1999) and that the loss of unc-26 would not decrease IP3 and DAG levels (Figure 3B), our result suggests that accumulation of membrane PIP2 suppresses the defects of ttx-7 mutants.
Figure 4.
A mutation in the unc-26 gene suppresses the synaptic defects of ttx-7 mutants. (A–C) The localization indices of SNB-1::VENUS (A), GFP::SYD-2 (B), and GLR-1::GFP (C). A mutation in unc-26 significantly suppressed the localization defects of the synaptic proteins in ttx-7(nj50) mutants. The marks on the bars of each genotype indicate comparisons with wild type. The marks on the lines indicate comparisons between indicated genotypes. Steel–Dwass multiple comparison tests were performed (n ≥ 11 animals). (D–F) Representative confocal images of the RIA neuron expressing SNB-1::VENUS (D), GFP::SYD-2 (E), and GLR-1::GFP (F) in ttx-7(nj50);unc-26(e205) double mutants. Scale bar, 5 µm. (G–I) Schematics of SNB-1::VENUS (G), GFP::SYD-2 (H), GLR-1::GFP (I) localizations in the RIA neuron of ttx-7(nj50);unc-26(e205) double mutants.
The ppk-1 gene encodes a type I PIP kinase that is regarded as a primary synthetic enzyme for PIP2 in vivo (Figure 3B; Stephens et al. 1991; Whiteford et al. 1997; Weinkove et al. 2008). Weinkove et al. (2008) showed that the overexpression of ppk-1 under a panneural promoter significantly increases PIP2 levels, while ppk-1(ok1141) mutants display an early larval lethal phenotype (Weinkove et al. 2008). We examined whether the overexpression of ppk-1 suppresses the defects of ttx-7 mutants. SNB-1::VENUS was still mislocalized in the nonpresynaptic region of RIA in the ppk-1 overexpression strain with ttx-7(nj50) mutation, as observed in ttx-7(nj50) single-mutant animals (Figure S4C and Table 1). The ppk-2 and ppk-3 genes encode homologs of type II and III PIP kinase, respectively. Type II kinase generates PIP2, and type III kinase generates phosphatidylinositol 3,5-bisphosphate (Figure 3B; Nicot et al. 2006; Weinkove et al. 2008). In both mutants, SNB-1::VENUS was normally localized at the presynaptic region (Table 1). In addition, we examined mutants for genes encoding phospholipase D (pld-1), ADP-ribosylation factor (arf-1.1, arf-1.2, arf-6), and Lowe oculocerebrorenal syndrome protein (ocrl-1). Homologs of these genes are involved in the metabolism of PIP2 in the Golgi apparatus in mammals (De Matteis et al. 2002; Di Paolo and De Camilli 2006). We did not identify any defects in these mutants (Table 1). We also tested mutations in the genes encoding phospholipase C isozymes (plc-1, plc-2) and proteins with the PI kinase domain (age-1, piki-1, vps-1, F35H12.4, Y75B8A.24, trr-1, smg-1, atm-1, atl-1, let-363), but these mutations neither caused a ttx-7-like defect nor suppressed the defect of ttx-7 mutants (Table 1). These results suggest that TTX-7 and the PI-related enzymes examined here function in distinct PI metabolic processes.
The synaptic defect in ttx-7 mutants is not merely a reflection of any known defects of polarity genes
The synaptic defects in ttx-7 mutants might be caused by defects in a selective transport system of synaptic proteins. We tested this possibility with a mutation in LRK-1, a homolog of familial parkinsonism gene PARK8/LRRK2, which causes a defect in the selective transport system, resulting in the abnormal localization of SNB-1 in sensory neurons (Sakaguchi-Nakashima et al. 2007). We did not, however, find any mislocalization defects of SNB-1 in the RIA neuron of lrk-1(km17) mutants (Table 1). Although the localization defect of SNB-1 in sensory neurons of lrk-1 mutants is suppressed by the loss of UNC-101 that is required for the transport of postsynaptic proteins (Sakaguchi-Nakashima et al. 2007), the mislocalization defect of SNB-1::VENUS in RIA neuron of ttx-7 mutants was not suppressed by a null mutation in unc-101 (Table 1; Dwyer et al. 2001). These results suggest that the molecular mechanism for the polarized localization of SNB-1 in RIA is different from that in sensory neurons.
Another possibility for the mechanism of the mislocalization is that the physical barriers between subcellular compartments in RIA are broken in ttx-7 mutants. A recent study revealed that ankyrin G is necessary for the cytosolic filter of axon initial segments (Song et al. 2009). We found that mutants for unc-44 gene encoding a C. elegans homolog of ankyrin G (Otsuka et al. 1995) showed normal localization of SNB-1::VENUS in RIA (Table 1).
Studies of cultured neurons have identified several molecules needed for cell polarity (Arimura and Kaibuchi 2007; Takenawa and Suetsugu 2007). Of C. elegans homologs of these molecules, mutants for cell division control protein 42 (cdc-42) (Gotta et al. 2001; Kay and Hunter 2001), collapsin response mediator protein-2 (CRMP-2) (unc-33) (Tsuboi et al. 2005), neural Wiskott–Aldrich syndrome protein (N-WASP) (wsp-1), WASP family Verprolin-homologous protein (WAVE) (wve-1), and proline-rich WASP-interacting protein (WIP) (wip-1) (Sawa et al. 2003; Sawa and Takenawa 2006) all showed normal localization of SNB-1::VENUS in the RIA neuron (Table 1). These results suggest that the polarized localization of synaptic molecules in RIA is controlled by a novel mechanism for neuronal polarity.
Mislocalization of synaptic vesicle proteins occurs independently of UNC-104, a kinesin-like protein
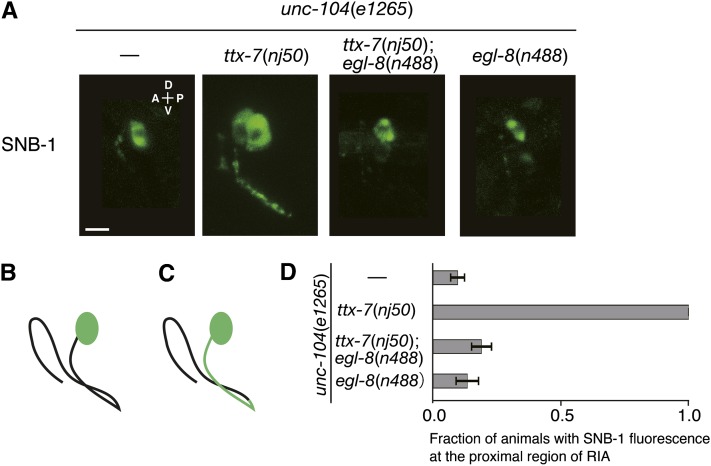
Kinesin motor proteins carry motor-specific cargos, assuring selective transport in neurons. UNC-104 is a kinesin-like motor protein and transports synaptic vesicles (SV) in C. elegans (Hall and Hedgecock 1991; Otsuka et al. 1991). Consistently, the fluorescence of SNB-1::VENUS was observed exclusively in the cell body of RIA in most of unc-104(e1265) mutant animals (Figure 5, A, B, and D). We examined whether the mislocalized SNB-1::VENUS in ttx-7 mutants is transported by UNC-104. We observed punctate fluorescence in the proximal region of the process in ttx-7(nj50);unc-104(e1265) double mutants, although the presynaptic localization of SNB-1::VENUS was abolished (Figure 5, A, C, and D). Further, this abnormal localization of SNB-1::VENUS was suppressed by egl-8(n488) (Figure 5, A, B, and D). These results suggest that some portion of SNB-1::VENUS is mislocalized by an UNC-104-independent pathway in ttx-7 mutants. Nonet et al. (1999) reported that a mutation in unc-11 causes diffused localization of SNB-1 in the nerve processes in a unc-104 background (Nonet et al. 1999). We found that the appearance of the SNB-1 fluorescence in unc-11(e47);unc-104(e1265) mutants was different from that of ttx-7(nj50);unc-104(e1265) mutants (Figure 5A and Figure S4B), suggesting that the losses of UNC-11 and TTX-7 cause mislocalization of SNB-1 in different processes.
Figure 5.
Localization of SNB-1 in a unc-104-mutant background. (A) Representative images of SNB-1::VENUS localization in the RIA neuron in each genotype. The fluorescence of SNB-1::VENUS in unc-104(e1265), ttx-7(nj50);unc-104(e1265);egl-8(n488), and unc-104(e1265);egl-8(n488) mutants was localized exclusively in the cell body, while it was abnormally localized as punctate in proximal region of the process in ttx-7(nj50);unc-104(e1265) mutants. Scale bar, 5 µm. (B and C) Schematic of SNB-1::VENUS localization in the RIA neuron of unc-104(e1265), ttx-7(nj50);egl-8(n488);unc-104(e1265), and egl-8(n488);unc-104(e1265) mutants (B) and that of ttx-7(nj50);unc-104(e1265) mutants (C). (D) The fraction of animals that displayed abnormal localization of SNB-1::VENUS in the proximal region of the RIA process in each genotype. SNB-1::VENUS expressed from integrated array njIs9 was observed. About 20 animals were examined in more than three trials.
egl-8 mutants are resistant to LiCl treatment on synaptic and thermotaxis phenotypes
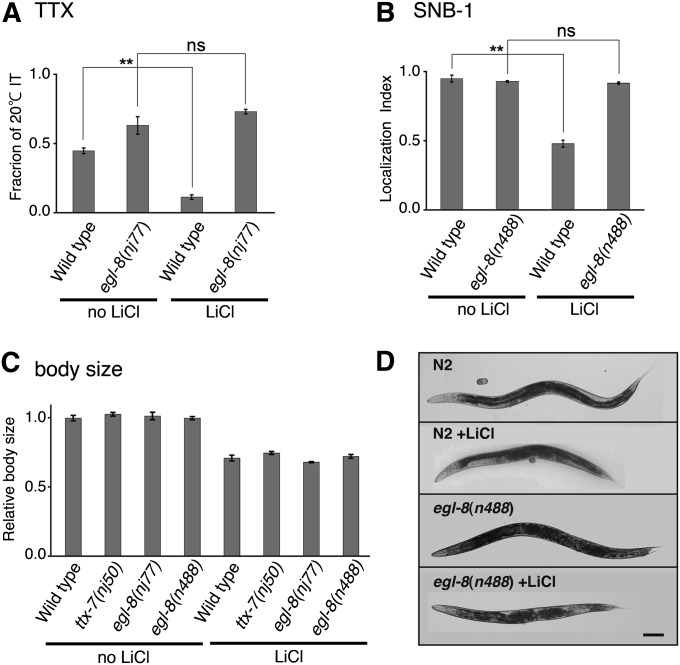
Lithium is used to treat bipolar disorder, and IMPase is one of the putative targets of lithium therapy (Hallcher and Sherman 1980; Berridge et al. 1989). Tanizawa et al. (2006) showed that the exogenous application of LiCl to wild-type animals mimics both the thermotaxis and synaptic defects of ttx-7 mutants (Tanizawa et al. 2006). Given the suppression of the ttx-7 defects by egl-8 mutations, we investigated whether egl-8 mutant animals are resistant to LiCl treatment. Treatment of wild-type animals with LiCl substantially reduced the fraction of IT behavior, while LiCl did not affect IT behavior of egl-8(nj77) mutants (Figure 6A). The localization of SNB-1::VENUS remained intact in LiCl-treated egl-8(n488) mutants unlike LiCl-treated wild-type animals (Figure 6B).
Figure 6.
Exogenous application of lithium to egl-8 mutants. (A) The effect of LiCl treatment on the individual thermotaxis assay in 20° cultivation. Lithium treatment significantly disrupted the IT behavior of wild-type animals, but not that of egl-8(nj77) mutants. About 20 animals were examined in more than three trials. Tukey–Kramer test was performed. (B) The localization indices of SNB-1::VENUS in the RIA neuron of animals treated with LiCl. SNB-1::VENUS in LiCl-treated wild-type animals was mislocalized to the proximal region of RIA, resulting in the lower localization index. egl-8(n488) mutants were completely resistant to lithium treatment. Steel–Dwass multiple comparison test was performed (n ≥ 12 animals). (C) Body size of lithium-treated and -untreated animals in each genotype relative to lithium-untreated wild-type animals. LiCl-treated animals were smaller than untreated animals. Mutations in egl-8 did not confer resistance in this case. n ≥ 19 animals. (D) Lateral views of lithium-treated and -untreated adult animals captured in bright fields. Scale bar, 100 µm.
We noted that LiCl treatment shortened the body length of animals (Figure 6, C and D). This is consistent with the previous reports that lithium interferes with normal development in various organisms (Gurvich and Klein 2002). The effect of LiCl on the body size does not appear to be caused by inhibition of IMPase, because the body size of ttx-7 mutants was normal and because LiCl treatment shortened the body size of ttx-7 mutants (Figure 6C). In contrast to the synaptic and thermotaxis phenotypes, mutations in egl-8 did not confer resistance to the developmental defect (Figure 6, C and D), indicating that resistance to LiCl conferred by the loss of EGL-8 function is specific to the abnormalities associated with IMPase dysfunction. These results suggest that lithium impairs synthesis of PIP2 through inhibition of IMPase, which causes the defect in the synaptic polarity of RIA.
Discussion
The PIP2 level in central interneurons of C. elegans is regulated by IMPase
Despite its well-known enzymatic property, the in vivo function of IMPase in neuronal PI metabolism has remained elusive. In this study, we show that the localization defects of synaptic proteins in IMPase ttx-7 mutants can be suppressed by disrupting PIP2 breakdown mediated by two membrane-associated enzymes, PLCβ EGL-8 or synaptojanin UNC-26. These results suggest that the neuronal PIP2 level is regulated by IMPase in vivo and that the proper PIP2 level is required for the synaptic polarity in a specific type of central neuron and thereby for normal behaviors.
We did not identify any other mutations in PI metabolic genes that suppress the defects of ttx-7 mutants or cause a defect in localization of synaptic proteins similar to that in ttx-7 mutants (Table 1). Further, the overexpression of ppk-1, which is known to be a major PIP2-producing enzyme (Weinkove et al. 2008), had no effect on the synaptic defects of ttx-7 mutants (Figure S4C and Table 1). These results suggest that TTX-7 and the PI metabolic enzymes tested here act in different domains of PI metabolism in RIA. Consistently, although PI is an important regulator of cell morphology (Skwarek and Boulianne 2009), the RIA morphology in ttx-7 mutants is almost normal (Figure 2F). Further, a certain amount of SV proteins are transported to the presynaptic region by the PIP2-dependent kinesin motor UNC-104 (Figure 2 and Figure 5; Klopfenstein and Vale 2004). These results suggest that the level of PI in RIA is not drastically reduced in ttx-7 mutants and that IMPase regulates a specialized part of the entire PI metabolism in the cell to localize synaptic molecules.
PI-mediated signaling regulates the polarized localization of pre- and postsynaptic components
How does the IMPase-mediated PI signaling localize synaptic molecules? Our study showed that some portion of the SV protein SNB-1 is mislocalized in ttx-7 mutants with a mutation in the kinesin SV transporter UNC-104 (Figure 5). Although careful interpretation is required because the unc-104 mutation is not null, this result suggests that the mislocalization of SNB-1 is not caused merely by the defect in the movement of the transporter. As mentioned by Nonet et al. (1999), a defect in the endocytosis process caused by a unc-11 mutation leads to mislocalization of SNB-1 along the membrane of neuronal processes in a unc-104 mutant background (Nonet et al. 1999). However, the smooth membranous appearance of SNB-1::VENUS in unc-11;unc-104 animals is different from the punctate appearance in ttx-7;unc-104 animals (Figure 5A and Figure S4B). Further, the mislocalization of SNB-1::VENUS in ttx-7;unc-104 was restricted to the proximal region of the process, while in unc-11;unc-104 animals, SNB-1::VENUS was visible across the entire process (Figure 5A and Figure S4B), suggesting that TTX-7 and UNC-11 are involved in distinct processes.
It is possible that UNC-11-independent endocytosis is defective in ttx-7 mutants. Margeta et al. (2009) showed that UNC-101, AP-1 medium subunit µ1, endocytically eliminates postsynaptic components at the presynaptic region of the RIA neuron (Margeta et al. 2009). Thus, we can speculate that another endocytotic machinery also functions in the PI signaling-dependent manner to eliminate presynaptic proteins such as SNB-1 at the postsynaptic region. We found that the localization of UNC-101 was disrupted in ttx-7 mutant animals (Figure S4A). This result implies that functional localization of such endocytotic machinery is also under the control of the PI signaling.
An alternative possibility is a defect of a selective transport system. Since many synaptic components contain the PIP2 binding domain, PIP2 might function as a signal for synaptic components to ride on specific cargos at the Golgi apparatus. However, given that EGL-8 and UNC-26 function on the cell membrane, it is likely that the loss of TTX-7 does not significantly affect the level of PIP2 at the Golgi apparatus.
A defect in cytoplasmic barriers might also cause the localization defects. Studies in vertebrates showed that ankyrin G- and F-actin are essential for cytoplasmic barriers to regulate protein localization in neurons (Nakada et al. 2003; Song et al. 2009). Although the loss of unc-44, which is the closest gene to ankyrin G in C. elegans, did not affect the localization of SNB-1::VENUS in RIA (Table 1), this result does not exclude the presence of such barriers.
The synaptic phenotype of ttx-7 mutants is unique: the disruption of all examined genes related to synapse formation or polarity establishment did not cause the ttx-7-like defects (Table 1; Tanizawa et al. 2006). Thus, it is likely that the PI signaling regulated by IMPase plays a fundamental role in a novel mechanism of synaptic localization. Andreassi et al. (2010) showed that the mRNA of mice IMPase accumulated in the axon of sympathetic neurons, also implying an important role of IMPase in neuronal processes (Andreassi et al. 2010). Although technically challenging, an electron microscopy analysis will be informative to fully understand how IMPase regulates localization of synaptic molecules.
Lithium impairs PIP2-mediated signaling through inhibition of IMPase in specific neurons
IMPase is a potent target of lithium therapy for bipolar disorder (Hallcher and Sherman 1980). It is hypothesized that the inhibition of IMPase by lithium reduces the inositol supply, which in turn interferes with a PIP2-mediated signaling pathway (Figure S2; Allison and Stewart 1971; Allison et al. 1976; Berridge et al. 1982, 1989; Schloesser et al. 2008). Indeed, a recent study using the social amoeba Dictyostelium showed that lithium treatment disrupts synthesis of PI species (King et al. 2009). However, inhibition of IMPase by lithium has never been shown to reduce neuronal PI levels in vivo, and studies suggesting the limited importance of IMPase in neuronal PI metabolism are accumulating (Godfrey et al. 1989; Batty and Downes 1994; Dixon et al. 1994; Schloesser et al. 2008; O’brien and Klein 2009). Of those, a study with mice lacking the SMIT1 gene that is required for taking up inositol from the extracellular environment (Figure 3B; Berry et al. 2004) showed that the loss of SMIT1 causes 92% reduction of inositol in fetal brain but does not affect PI levels. This result implies that the low concentration of inositol is sufficient to synthesize PI species. Considering that the reduction of inositol by IMPase inhibition is much more modest compared with that in the SMIT1 knockout mice, the authors claimed that the inositol depletion hypothesis is not probable (Berry et al. 2004). However, if the inhibition of IMPase reduces PI levels in a specific region of the nervous system and a unique metabolic module within a cell, the global measurement would not reveal the reduction of PI levels. As shown by Tanizawa et al. (2006), the localization defects of synaptic proteins in ttx-7 mutants occur exclusively in the RIA neuron (Tanizawa et al. 2006). Further, this study showed that IMPase is involved in a specialized part of PI metabolism in the cell. Thus, inhibition of IMPase in human brain might also affect only specific types of neurons and also specific types of metabolic modules of PI metabolism in the neurons. This idea can help explain the controversial results obtained by different experimental samples.
The RIA neuron has numerous synapses in its single neurite. The high level of total synaptic activity might consume a large amount of inositol, making RIA sensitive to the IMPase inhibition. If so, neurons with a relatively large number of synapses can be a potent candidate for the target of lithium treatment in human brain.
It was shown that knockout of the IMPA1 or IMPA2 genes encoding IMPase in mice does not decrease the global level of inositol in the adult brain (Cryns et al. 2007, 2008; Agam et al. 2009), which is consistent with the result that ttx-7 mutants does not show any defects related to the known inositol and PI-mediated signaling (Tanizawa et al. 2006). It is plausible that mammalian IMPase also acts in a specialized part of PI metabolism.
We also found that synaptojanin UNC-26 is linked to the PI metabolism in which IMPase and PLCβ function. Genetic studies on human patients suggested that synaptojanin 1 is also associated with bipolar disorder (Saito et al. 2001; Stopkova et al. 2004), raising an intriguing hypothesis that the metabolic module of IMPase, PLCβ, and synaptojanin at synapses is a site of lithium action.
Supplementary Material
Acknowledgments
We thank M. Nonet for pSB120; C. Rongo for CR120; M. Zhen for pJH23; P. Sengupta for ofm-1p::gfp; S. Nurrish for REW1 and KP316; K. Kimura for pKDK66; D. Weinkove for EG3361; N. Hisamoto and K. Matsumoto for lrk-1(km17) and pld-1(km22) mutant strains; Caenorhabditis Genetic Center and National Bioresource Project (Japan) for strains; M. Okumura for the glr-3 promoter; A. Fire for pPD plasmid; and the members of the Mori laboratory for fruitful discussions. T.K. was supported by the Japan Society for the Promotion of Science. This work was supported by CREST, Japan Science and Technology Agency, and Grant-in-Aid for Scientific Research on Innovative Areas “Neural Diversity and Neocortical Organization” from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan to IM.
Footnotes
Communicating editor: K. Kemphues
Literature Cited
- Agam G., Bersudsky Y., Berry G. T., Moechars D., Lavi-Avnon Y., et al. , 2009. Knockout mice in understanding the mechanism of action of lithium. Biochem. Soc. Trans. 37: 1121–1125 [DOI] [PubMed] [Google Scholar]
- Allison J. H., Stewart M. A., 1971. Reduced brain inositol in lithium-treated rats. Nat. New Biol. 233: 267–268 [DOI] [PubMed] [Google Scholar]
- Allison J. H., Blisner M. E., Holland W. H., Hipps P. P., Sherman W. R., 1976. Increased brain myo-inositol 1-phosphate in lithium-treated rats. Biochem. Biophys. Res. Commun. 71: 664–670 [DOI] [PubMed] [Google Scholar]
- Andreassi C., Zimmermann C., Mitter R., Fusco S., De Vita S., et al. , 2010. An NGF-responsive element targets myo-inositol monophosphatase-1 mRNA to sympathetic neuron axons. Nat. Neurosci. 13: 291–301 [DOI] [PubMed] [Google Scholar]
- Arimura N., Kaibuchi K., 2005. Key regulators in neuronal polarity. Neuron 48: 881–884 [DOI] [PubMed] [Google Scholar]
- Arimura N., Kaibuchi K., 2007. Neuronal polarity: from extracellular signals to intracellular mechanisms. Nat. Rev. Neurosci. 8: 194–205 [DOI] [PubMed] [Google Scholar]
- Batty I. H., Downes C. P., 1994. The inhibition of phosphoinositide synthesis and muscarinic-receptor-mediated phospholipase C activity by Li+ as secondary, selective, consequences of inositol depletion in 1321N1 cells. Biochem. J. 297(3): 529–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge M. J., Downes C. P., Hanley M. R., 1982. Lithium amplifies agonist-dependent phosphatidylinositol responses in brain and salivary glands. Biochem. J. 206: 587–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge M. J., Downes C. P., Hanley M. R., 1989. Neural and developmental actions of lithium: a unifying hypothesis. Cell 59: 411–419 [DOI] [PubMed] [Google Scholar]
- Berry G. T., Buccafusca R., Greer J. J., Eccleston E., 2004. Phosphoinositide deficiency due to inositol depletion is not a mechanism of lithium action in brain. Mol. Genet. Metab. 82: 87–92 [DOI] [PubMed] [Google Scholar]
- Brenner S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cade J. F. J., 1949. Lithium salts in the treatment of psychotic excitement. Med. J. Aust. 2: 349–352 [DOI] [PubMed] [Google Scholar]
- Cremona O., Di Paolo G., Wenk M. R., Lüthi A., Kim W. T., et al. , 1999. Essential role of phosphoinositide metabolism in synaptic vesicle recycling. Cell 99: 179–188 [DOI] [PubMed] [Google Scholar]
- Cryns K., Shamir A., Shapiro J., Daneels G., Goris I., et al. , 2007. Lack of lithium-like behavioral and molecular effects in IMPA2 knockout mice. Neuropsychopharmacology 32: 881–891 [DOI] [PubMed] [Google Scholar]
- Cryns K., Shamir A., Van Acker N., Levi I., Daneels G., et al. , 2008. IMPA1 is essential for embryonic development and lithium-like pilocarpine sensitivity. Neuropsychopharmacology 33: 674–684 [DOI] [PubMed] [Google Scholar]
- De Matteis M., Godi A., Corda D., 2002. Phosphoinositides and the golgi complex. Curr. Opin. Cell Biol. 14: 434–447 [DOI] [PubMed] [Google Scholar]
- Di Paolo G., De Camilli P., 2006. Phosphoinositides in cell regulation and membrane dynamics. Nature 443: 651–657 [DOI] [PubMed] [Google Scholar]
- Dixon J. F., Los G. V., Hokin L. E., 1994. Lithium stimulates glutamate “release” and inositol 1,4,5-trisphosphate accumulation via activation of the N-methyl-d-aspartate receptor in monkey and mouse cerebral cortex slices. Proc. Natl. Acad. Sci. USA 91: 8358–8362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer N. D., Adler C. E., Crump J. G., L’Etoile N. D., Bargmann C. I., 2001. Polarized dendritic transport and the AP-1 mu1 clathrin adaptor UNC-101 localize odorant receptors to olfactory cilia. Neuron 31: 277–287 [DOI] [PubMed] [Google Scholar]
- Godfrey P. P., McClue S. J., White A. M., Wood A. J., Grahame-Smith D. G., 1989. Subacute and chronic in vivo lithium treatment inhibits agonist- and sodium fluoride-stimulated inositol phosphate production in rat cortex. J. Neurochem. 52: 498–506 [DOI] [PubMed] [Google Scholar]
- Gotta M., Abraham M. C., Ahringer J., 2001. CDC-42 controls early cell polarity and spindle orientation in C. elegans. Curr. Biol. 11: 482–488 [DOI] [PubMed] [Google Scholar]
- Gurvich N., Klein P. S., 2002. Lithium and valproic acid: parallels and contrasts in diverse signaling contexts. Pharmacol. Ther. 96: 45–66 [DOI] [PubMed] [Google Scholar]
- Hall D. H., Hedgecock E. M., 1991. Kinesin-related gene unc-104 is required for axonal transport of synaptic vesicles in C. elegans. Cell 65: 837–847 [DOI] [PubMed] [Google Scholar]
- Hallcher L. M., Sherman W. R., 1980. The effects of lithium ion and other agents on the activity of myo-inositol-1-phosphatase from bovine brain. J. Biol. Chem. 255: 10896–10901 [PubMed] [Google Scholar]
- Harris T. W., Hartwieg E., Horvitz H. R., Jorgensen E. M., 2000. Mutations in synaptojanin disrupt synaptic vesicle recycling. J. Cell Biol. 150: 589–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgecock E. M., Russell R. L., 1975. Normal and mutant thermotaxis in the nematode Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 72: 4061–4065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedgepeth C. M., Conrad L. J., Zhang J., Huang H. C., Lee V. M., et al. , 1997. Activation of the Wnt signaling pathway: a molecular mechanism for lithium action. Dev. Biol. 185: 82–91 [DOI] [PubMed] [Google Scholar]
- Ito H., Inada H., Mori I., 2006. Quantitative analysis of thermotaxis in the nematode Caenorhabditis elegans. J. Neurosci. Methods 154: 45–52 [DOI] [PubMed] [Google Scholar]
- Jurado P., Kodama E., Tanizawa Y., Mori I., 2010. Distinct thermal migration behaviors in response to different thermal gradients in Caenorhabditis elegans. Genes Brain Behav. 9: 120–127 [DOI] [PubMed] [Google Scholar]
- Kay A. J., Hunter C. P., 2001. CDC-42 regulates PAR protein localization and function to control cellular and embryonic polarity in C. elegans. Curr. Biol. 11: 474–481 [DOI] [PubMed] [Google Scholar]
- King J. S., Teo R., Ryves J., Reddy J. V., Peters O., et al. , 2009. The mood stabiliser lithium suppresses PIP3 signalling in Dictyostelium and human cells. Dis. Model Mech. 2: 306–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopfenstein D. R., Vale R. D., 2004. The lipid binding pleckstrin homology domain in UNC-104 kinesin is necessary for synaptic vesicle transport in Caenorhabditis elegans. Mol. Biol. Cell 15: 3729–3739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu H., Mori I., Rhee J. S., Akaike N., Ohshima Y., 1996. Mutations in a cyclic nucleotide-gated channel lead to abnormal thermosensation and chemosensation in C. elegans. Neuron 17: 707–718 [DOI] [PubMed] [Google Scholar]
- Lackner M. R., Nurrish S. J., Kaplan J. M., 1999. Facilitation of synaptic transmission by EGL-30 Gqalpha and EGL-8 PLCbeta: DAG binding to UNC-13 is required to stimulate acetylcholine release. Neuron 24: 335–346 [DOI] [PubMed] [Google Scholar]
- Lee Y.-S., Mulugu S., York J. D., O’Shea E. K., 2007. Regulation of a cyclin-CDK-CDK inhibitor complex by inositol pyrophosphates. Science 316: 109–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L., Clark D. A., Biron D., Mahadevan L., Samuel A. D. T., 2006. Sensorimotor control during isothermal tracking in Caenorhabditis elegans. J. Exp. Biol. 209: 4652–4662 [DOI] [PubMed] [Google Scholar]
- Machado-Vieira R., Manji H. K., Zarate J., Carlos A., 2009. The role of lithium in the treatment of bipolar disorder: convergent evidence for neurotrophic effects as a unifying hypothesis. Bipolar Disord. 11(Suppl. 2): 92–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margeta M. A., Wang G. J., Shen K., 2009. Clathrin adaptor AP-1 complex excludes multiple postsynaptic receptors from axons in C. elegans. Proc. Natl. Acad. Sci. USA 106: 1632–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslanski J. A., Leshko L., Busa W. B., 1992. Lithium-sensitive production of inositol phosphates during amphibian embryonic mesoderm induction. Science 256: 243–245 [DOI] [PubMed] [Google Scholar]
- Mello C., Kramer J., Stinchcomb D., Ambros V., 1991. Efficient gene transfer in C. elegans: extrachromosomal maintenance and inte-gration of transforming sequences. EMBO J. 10: 3959–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K. G., Emerson M. D., Rand J. B., 1999. Goalpha and diacylglycerol kinase negatively regulate the Gqalpha pathway in C. elegans. Neuron 24: 323–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohri A., Kodama E., Kimura K. D., Koike M., Mizuno T., et al. , 2005. Genetic control of temperature preference in the nematode Caenorhabditis elegans. Genetics 169: 1437–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori I., Ohshima Y., 1995. Neural regulation of thermotaxis in Caenorhabditis elegans. Nature 376: 344–348 [DOI] [PubMed] [Google Scholar]
- Nakada C., Ritchie K., Oba Y., Nakamura M., Hotta Y., et al. , 2003. Accumulation of anchored proteins forms membrane diffusion barriers during neuronal polarization. Nat. Cell Biol. 5: 626–632 [DOI] [PubMed] [Google Scholar]
- Nicot A.-S., Fares H., Payrastre B., Chisholm A. D., Labouesse M., et al. , 2006. The phosphoinositide kinase PIKfyve/Fab1p regulates terminal lysosome maturation in Caenorhabditis elegans. Mol. Biol. Cell 17: 3062–3074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet M. L., Holgado A. M., Brewer F., Serpe C. J., Norbeck B. A., et al. , 1999. UNC-11, a Caenorhabditis elegans AP180 homologue, regulates the size and protein composition of synaptic vesicles. Mol. Biol. Cell 10: 2343–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien W. T., Klein P. S., 2009. Validating GSK3 as an in vivo target of lithium action. Biochem. Soc. Trans. 37: 1133–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell T., Rotzinger S., Nakashima T. T., Hanstock C. C., Ulrich M., et al. , 2000. Chronic lithium and sodium valproate both decrease the concentration of myo-inositol and increase the concentration of inositol monophosphates in rat brain. Brain Res. 880: 84–91 [DOI] [PubMed] [Google Scholar]
- Odom A. R., Stahlberg A., Wente S. R., York J. D., 2000. A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science 287: 2026–2029 [DOI] [PubMed] [Google Scholar]
- Ohnishi T., Watanabe A., Ohba H., Iwayama Y., Maekawa M., et al. , 2010. Behavioral analyses of transgenic mice harboring bipolar disorder candidate genes, IMPA1 and IMPA2. Neurosci. Res. 67: 86–94 [DOI] [PubMed] [Google Scholar]
- Okochi Y., Kimura K. D., Ohta A., Mori I., 2005. Diverse regulation of sensory signaling by C. elegans nPKC-epsilon/eta TTX-4. EMBO J. 24: 2127–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka A. J., Jeyaprakash A., Garcia-Anoveros J., Tang L. Z., Fisk G., et al. , 1991. The C. elegans unc-104 gene encodes a putative kinesin heavy chain-like protein. Neuron 6: 113–122 [DOI] [PubMed] [Google Scholar]
- Otsuka A. J., Franco R., Yang B., Shim K. H., Tang L. Z., et al. , 1995. An ankyrin-related gene (unc-44) is necessary for proper axonal guidance in Caenorhabditis elegans. J. Cell Biol. 129: 1081–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu W. S., Samuel A. D. T., 2002. Thermotaxis in Caenorhabditis elegans analyzed by measuring responses to defined thermal stimuli. J. Neurosci. 22: 5727–5733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T., Guan F., Papolos D. F., Lau S., Klein M., et al. , 2001. Mutation analysis of SYNJ1: a possible candidate gene for chromosome 21q22-linked bipolar disorder. Mol. Psychiatry 6: 387–395 [DOI] [PubMed] [Google Scholar]
- Sakaguchi-Nakashima A., Meir J. Y., Jin Y., Matsumoto K., Hisamoto N., 2007. LRK-1, a C. elegans PARK8-related kinase, regulates axonal-dendritic polarity of SV proteins. Curr. Biol. 17: 592–598 [DOI] [PubMed] [Google Scholar]
- Sawa M., Takenawa T., 2006. Caenorhabditis elegans WASP-interacting protein homologue WIP-1 is involved in morphogenesis through maintenance of WSP-1 protein levels. Biochem. Biophys. Res. Commun. 340: 709–717 [DOI] [PubMed] [Google Scholar]
- Sawa M., Suetsugu S., Sugimoto A., Miki H., Yamamoto M., et al. , 2003. Essential role of the C. elegans Arp2/3 complex in cell migration during ventral enclosure. J. Cell Sci. 116: 1505–1518 [DOI] [PubMed] [Google Scholar]
- Schloesser R. J., Huang J., Klein P. S., Manji H. K., 2008. Cellular plasticity cascades in the pathophysiology and treatment of bipolar disorder. Neuropsychopharmacology 33: 110–133 [DOI] [PubMed] [Google Scholar]
- Seeds A. M., Bastidas R. J., York J. D., 2005. Molecular definition of a novel inositol polyphosphate metabolic pathway initiated by inositol 1,4,5-trisphosphate 3-kinase activity in Saccharomyces cerevisiae. J. Biol. Chem. 280: 27654–27661 [DOI] [PubMed] [Google Scholar]
- Shaldubina A., Ju S., Vaden D. L., Ding D., Belmaker R. H., et al. , 2002. Epi-inositol regulates expression of the yeast INO1 gene encoding inositol-1-P synthase. Mol. Psychiatry 7: 174–180 [DOI] [PubMed] [Google Scholar]
- Skwarek L. C., Boulianne G. L., 2009. Great expectations for PIP: phosphoinositides as regulators of signaling during development and disease. Dev. Cell 16: 12–20 [DOI] [PubMed] [Google Scholar]
- Song A.-H., Wang D., Chen G., Li Y., Luo J., et al. , 2009. A selective filter for cytoplasmic transport at the axon initial segment. Cell 136: 1148–1160 [DOI] [PubMed] [Google Scholar]
- Stephens L. R., Hughes K. T., Irvine R. F., 1991. Pathway of phosphatidylinositol(3,4,5)-trisphosphate synthesis in activated neutrophils. Nature 351: 33–39 [DOI] [PubMed] [Google Scholar]
- Stopkova P., Vevera J., Paclt I., Zukov I., Lachman H. M., 2004. Analysis of SYNJ1, a candidate gene for 21q22 linked bipolar disorder: a replication study. Psychiatry Res. 127: 157–161 [DOI] [PubMed] [Google Scholar]
- Takenawa T., Suetsugu S., 2007. The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat. Rev. Mol. Cell Biol. 8: 37–48 [DOI] [PubMed] [Google Scholar]
- Tanizawa Y., Kuhara A., Inada H., Kodama E., Mizuno T., et al. , 2006. Inositol monophosphatase regulates localization of synaptic components and behavior in the mature nervous system of C. elegans. Genes Dev. 20: 3296–3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi D., Hikita T., Qadota H., Amano M., Kaibuchi K., 2005. Regulatory machinery of UNC-33 Ce-CRMP localization in neurites during neuronal development in Caenorhabditis elegans. J. Neurochem. 95: 1629–1641 [DOI] [PubMed] [Google Scholar]
- Weinkove D., Bastiani M., Chessa T. A. M., Joshi D., Hauth L., et al. , 2008. Overexpression of PPK-1, the Caenorhabditis elegans Type I PIP kinase, inhibits growth cone collapse in the developing nervous system and causes axonal degeneration in adults. Dev. Biol. 313: 384–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J. G., Southgate E., Thomson J. N., Brenner S., 1986. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 314: 1–340 [DOI] [PubMed] [Google Scholar]
- Whiteford C. C., Brearley C. A., Ulug E. T., 1997. Phosphatidylinositol 3,5-bisphosphate defines a novel PI 3-kinase pathway in resting mouse fibroblasts. Biochem. J. 323(3): 597–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicks S. R., Yeh R. T., Gish W. R., Waterston R. H., Plasterk R. H., 2001. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 28: 160–164 [DOI] [PubMed] [Google Scholar]
- Yeh E., Kawano T., Weimer R. M., Bessereau J.-L., Zhen M., 2005. Identification of genes involved in synaptogenesis using a fluorescent active zone marker in Caenorhabditis elegans. J. Neurosci. 25: 3833–3841 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.