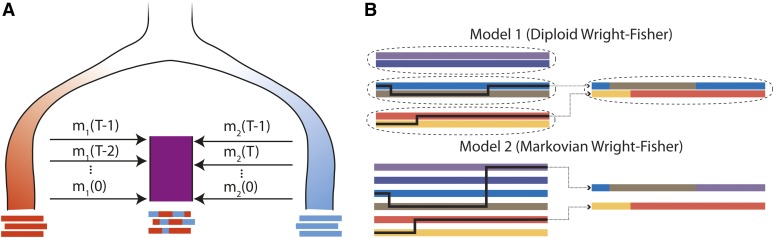
Figure 2 .
(A) Illustration of an admixture model starting at generation T − 1, where the admixed population (purple) receiving mi(t) migrants from diverged red (i = 1) and blue (i = 2) source populations at generation t. If these are statistically distinct enough, it is possible to infer the ancestry along the admixed chromosomes. Independent of our statistical power to infer this detailed local ancestry, the mosaic pattern may leave distinct traces in genome-wide statistics, such as global ancestry or linkage patterns. (B) Gamete formation in two versions of the Wright–Fisher model with recombination. In model 1, diploid individuals are generated by randomly selecting two parents and generating gametes by following a Markov paths along the parental chromosomes. In model 2, gametes are generated by following a Markovian path across the parental allele pool. Both models have the same distribution of crossover numbers and are equivalent for genomic regions small enough that multiple crossovers are unlikely. Model 1 is more biologically realistic and is used in the simulations, whereas model 2 is more tractable and is used for inference and analytic derivations.