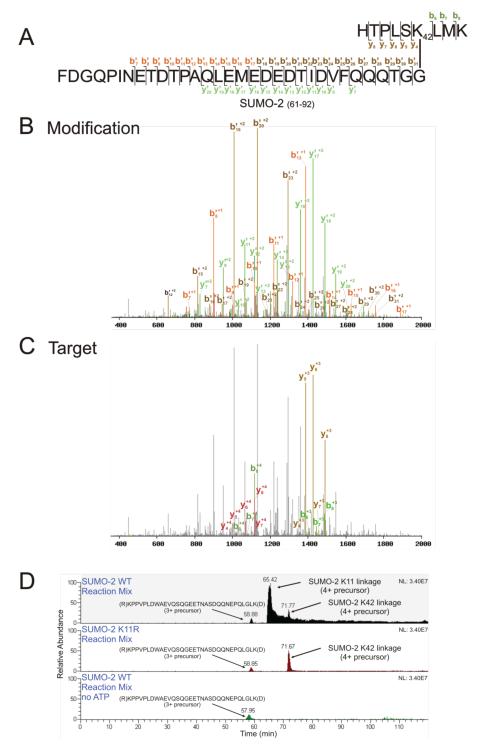
Figure 2.
K42-linked SUMO-2 is identified by SUMmOn. (A) Amino acid sequence of the trypsin-digested K42-linked SUMO-2-SUMO-2 peptide, indicating the ion fragments assigned by SUMmOn. (B) SUMmOn-annotated CID spectrum of the K42-linked SUMO-2-SUMO-2 peptide highlighting b’- and y’-ions derived from the modification (i.e. the C-terminal tryptic peptide of SUMO-2, aa 61-92). (C) The same spectrum, highlighting the b- and y-ions derived from the target peptide (aa 37-45 of SUMO-2). (D) Extracted ion chromatograms (EIC) of three different representative SUMO-2 in vitro reaction mixes. The m/z corresponding to the SUMO-2 K11 and K42 linkages elute at ~66 and ~72 min, respectively (top panel). The m/z corresponding to the SUMO-2 K11 linkage is lost when a K11R SUMO-2 mutant protein is used in the in vitro conjugation reaction, while the K42-linked peptide is unaffected (middle panel). The m/z corresponding to the K42 linkage is also lost in a reaction mix lacking ATP (bottom panel). Indicated are (i) the EIC of the SAE2 (SUMO E1) 3+ peptide (R)KPPVPLDWAEVQSQGEETNASDQQNEPQLGLK(D), eluting at ~ 59 min, using an m/z window of 1178.57-1178.59, (ii) an EIC of the 4+ (the most abundant charge state) SUMO-2 K11 linked peptide, using an m/z window of 1291.59-1291.61, and (iii) the EIC of the 4+ SUMO-2 K42 linked peptide (the most abundant charge state), using an m/z window of 1152.29-1152.31. For very large peptides, such as those characterized here, the monoisotopic variant represents only a very small percentage of the peptide population. The m/z windows in Figures 2, 4 and 7 therefore do not bracket the monoisotopic m/z for each linkage, but instead encompass the most abundant isotopic variants (i.e. those that were most often isolated for fragmentation).