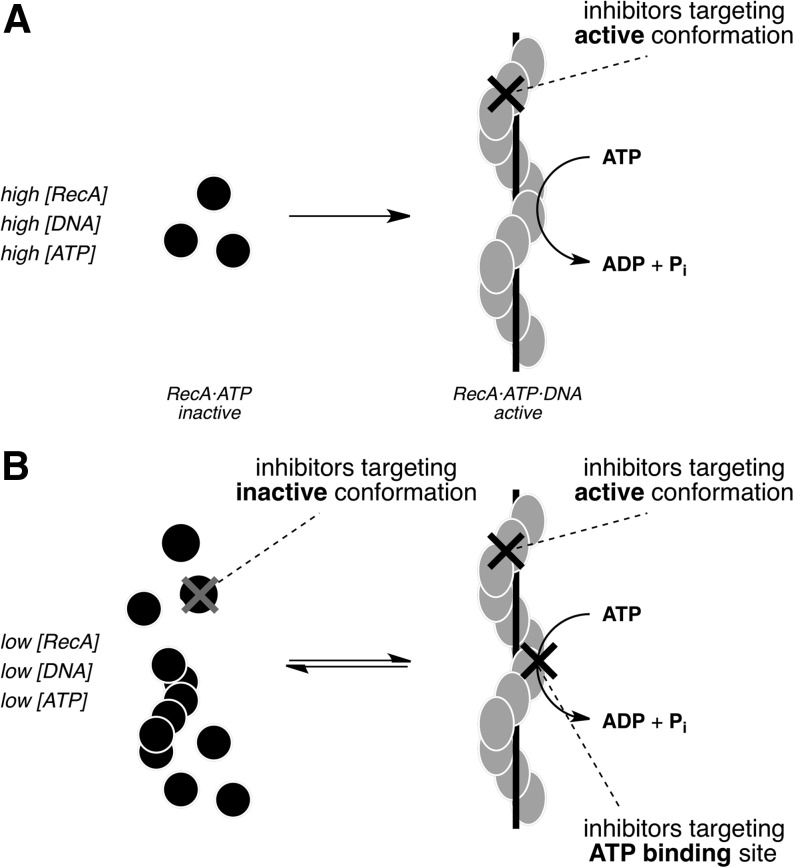
Fig. 1.
Cartoons depicting the inactive RecA monomers and active RecA-DNA filament (RDF). (A) Assay conditions, including high concentrations of RecA, DNA, and ATP, shift the filament assembly and activation equilibrium such that RecA exists almost exclusively in an active, DNA-bound conformational state. Inhibitors identified under such forcing conditions will be strongly biased toward binders of this conformational state. (B) The desired assay conditions include 10-fold lower concentrations of RecA and ATP, and RecA samples both its inactive and active conformational states during the assay. Such nonforcing conditions would facilitate the identification of additional inhibitors without bias with respect to RecA conformational preference as well as ATP-competitive inhibitors. ATP; adenosine 5′-O-triphosphate.