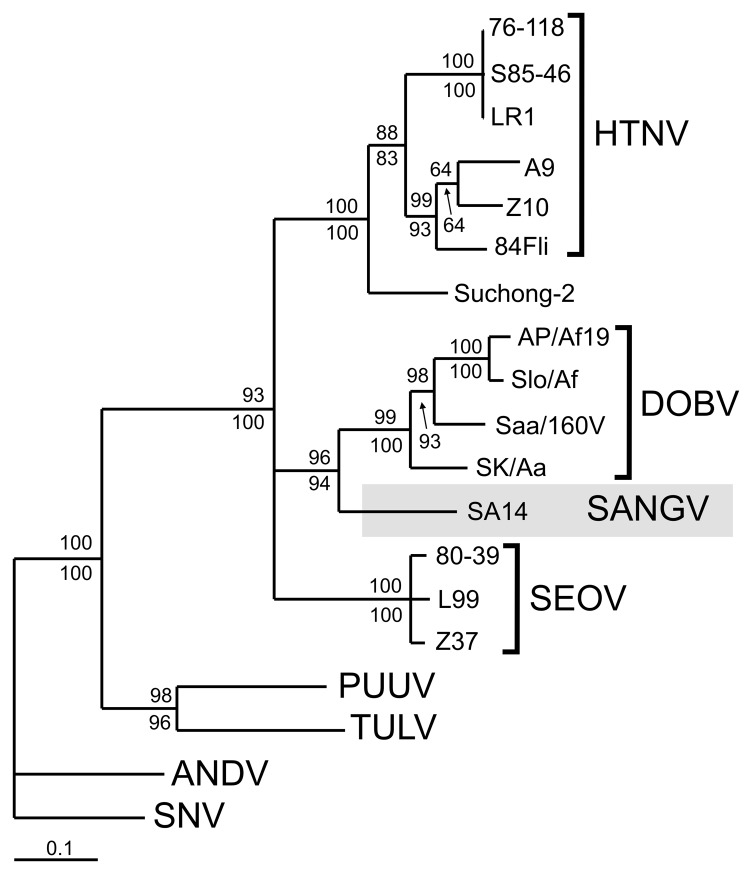
Figure.
Maximum likelihood phylogenetic tree of hantaviruses showing the placement of SA14 (Sangassou virus [SANGV], indicated by gray shading). Partial S segment genome sequences (837 nucleotides, positions 394–1230) were used to calculate the tree with TREE-PUZZLE program (8). The Tamura-Nei evolutionary model was used; the values above the branches represent PUZZLE support values. The values below the branches represent bootstrap values of the corresponding neighbor-joining tree computed with PAUP* program (9) using 10,000 bootstrap replicates. The scale bar indicates an evolutionary distance of 0.1 nucleotide substitutions per position in the sequence. HTNV, Hantaan virus; DOBV, Dobrava virus; SEOV, Seoul virus; PUUV, Puumala virus; TULV, Tula virus; ANDV, Andes virus; SNV, Sin Nombre virus.