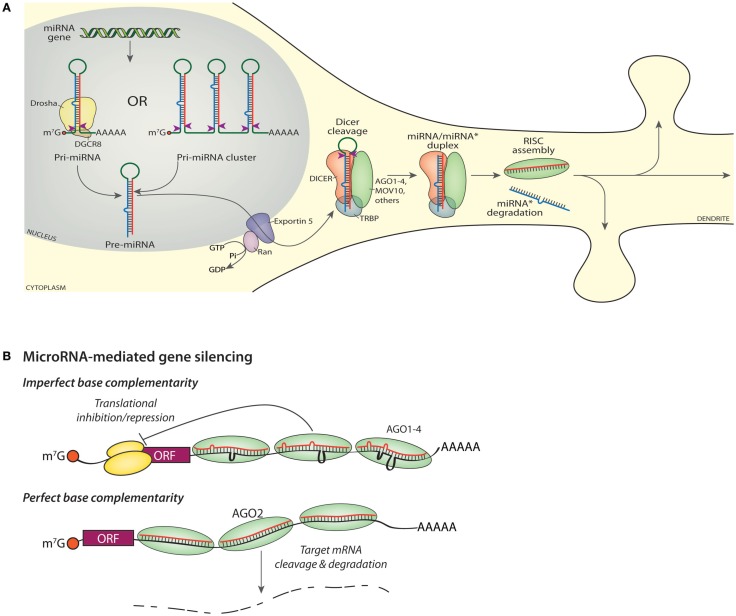
Figure 2.
miRNA biogenesis and gene silencing. (A) Transcription of a miRNA gene yields a capped, polyadenylated Pri-miRNA shown undergoing cleavage (red arrowheads) by Drosha (top, left). Transcription of a miRNA cluster yields a transcript encoding multiple miRNAs (top, right), each of which must be cleaved into a Pre-miRNA by Drosha. The mature or “guide” miRNA is shown in red, with the “star” or “passenger” strand shown in blue. Nuclear export of the Pre-miRNA requires Exportin-5. Cleavage of the Pre-miRNA by DICER (red arrowheads) generates a mature guide and star strand duplex; cleavage occurs in a protein complex that includes DICER, an Argonaute protein (AGO1-4), MOV10, TRBP, and additional proteins such as FMR1, DDX9, PUM2 (Schratt, 2009b; Czech and Hannon, 2011; De and MacRae, 2011). The star strand is lost from the final RISC and ultimately degraded, while the assembled RISC is available to bind to cognate RNAs and modulate protein expression. (B) Imperfect base complementarity. When the sequence of the miRNA guide strand does not perfectly match the target mRNA, translation of that mRNA is inhibited. Bound RISCs may contain any one of the four Argonaute proteins. The orange ball is the cap-binding complex; the large and small ribosomal subunits are shown in yellow. Perfect base complementarity. When the sequence of the miRNA guide strand matches the target mRNA perfectly, the target mRNA is sliced by AGO2.