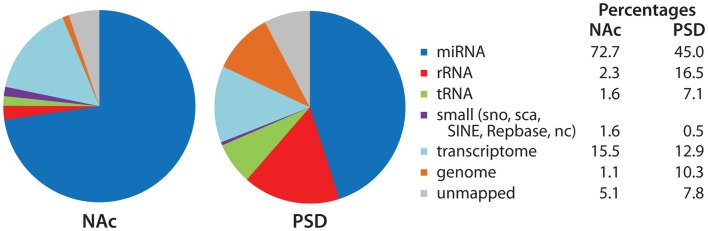
Figure 5.
Identity of mapped miRNA sequences in NAc and PSD libraries. Adaptor sequences were trimmed from the extracted sequence data. Bowtie (version 0.12.7; Langmead et al., 2009) was used to align sequences 18–30 nt in length to the miRBase annotated mouse microRNA database (version 16; Griffiths-Jones et al., 2006, 2008); perfect alignment was required. The Rfam, Repbase, and NON-CODE databases were used to identify sequences that aligned with annotated non-coding and repetitive elements. Sequences were then aligned to the mouse transcriptome (exons, introns, exon–exon junctions) and mouse genome. For non-miRNA alignments, 1-mismatch was allowed. Data (% total reads mapped to each category) for the NAc and PSD libraries from saline-injected animals are shown.