Abstract
Symbiosis between the bobtail squid Euprymna scolopes (Mollusca: Cephalopoda) and Vibrio fischeri bacteria has been a well-studied model for understanding the molecular mechanisms of colonization and adherence to host cells. For example, pilin expression has been observed to cause subtle variation in colonization for a number of Gram-negative bacteria with eukaryotic hosts. To investigate variation amongst pil genes of closely related strains of vibrios, we amplified pil genes A, B, C and D to determine orientation and sequence similarity to other symbiotic vibrios. The pilA gene was found to be upstream from all other pil genes, and not contiguous with the rest of the operon. The pilB, pilC and pilD loci were flanked at the 3′ end by yacE, followed by a conserved hypothetical gene. DNA sequences of each pil gene were aligned and analysed phylogenetically using parsimony for both individual and combined gene trees. Results demonstrate that certain pil loci (pilB and pilD) are conserved among strains of V. fischeri, but pilC differs in sequence between symbiotic and free-living strains. Phylogenetic analysis of all pil genes gives better resolution of Indo-west Pacific V. fischeri symbionts compared with analysis of the 16S rRNA gene. Hawaiian and Australian symbiotic strains form one monophyletic tree, supporting the hypothesis that V. fischeri strain specificity is selected by the geographical location of their hosts and is not related to specific squid species.
INTRODUCTION
The occurrence of bacterial symbionts with host partners is quite extensive and has been a topic for many evolutionary and developmental microbiologists (Nyholm & McFall-Ngai, 2004; Visick & Ruby, 2006). Vibrio fischeri belongs to the highly diverse family Vibrionaceae of the Gammaproteobacteria (Nishiguchi & Nair, 2003; Thompson et al., 2004a). These luminous bacteria specifically colonize the light organ of sepiolid squids and monocentrid fishes (McFall-Ngai, 1994; McFall-Ngai & Ruby, 1998; Jones & Nishiguchi, 2004). The association begins after hatching, where bacteria are acquired environmentally via the light organ pores, which are continuously exposed to surrounding seawater. Although all environmental bacteria are in contact with the pores, only specific strains of V. fischeri can colonize and persist therein, out-competing non-native strain even during direct competition (Nishiguchi et al., 1998; Nishiguchi, 2002).
Features known to play significant roles in success of the squid–symbiont relationship include chemotaxis, specific adherence, colonization, persistence and out-competing other bacterial strains (Graf et al., 1994; Millikan & Ruby, 2004; Nyholm & McFall-Ngai, 2004). Likewise, competition studies have determined that both ecological and environmental factors may be responsible for the evolutionary history of specificity between the partners (Nishiguchi et al., 1998; Nishiguchi, 2000, 2002). Although a number of biochemical and molecular mechanisms are found to increase the ability of V. fischeri to colonize and persist in the light organ, phenotypic characteristics such as pili have been observed to increase the surface area between bacterium and host (Nair, 2006). Pili are long, filamentous projections on the surface of both Gram-positive and Gram-negative bacteria, and are known to be important in both colonization and twitching motility (Darzins & Russell, 1997). They also mediate adherence or attachment to both abiotic and biotic surfaces (Stabb & Ruby, 2003). Pili are formed by a number of proteins encoded by genes found in the pil operon, and are grouped as types I, II, III and IV based on the pathway by which they are secreted (Soto & Hultgren, 1999). For example, V. fischeri expresses type IV pili, which are secreted by a pathway homologous to the type II secretion apparatus (Nunn, 1999; Sandkvist, 2001). The pilA gene codes for the major structural pilus component and pilB is involved in nucleotide binding, which controls the extension of type IV pili by the process of polymerization of the pilin subunit (Villar et al., 2001). pilC codes for a transmembrane protein that belongs to the GspF family of proteins and is involved in pilus assembly and adherence (Winther-Larsen et al., 2001), while pilD serves as a leader peptidase for processing PilA precursors and N-methylation of the first amino acid of the mature protein. Previous observations of different wild-type V. fischeri strains isolated from both seawater and squid light organs exhibited differences in pili (both in vitro and in vivo; Nair, 2006), which may influence the specificity between sepiolid squids and their Vibrio symbionts and may be useful loci to determine subtle strain differences when determining the phylogenetic relatedness between symbiotic and free-living strains of V. fischeri. Therefore, we sequenced individual pil genes from strains of V. fischeri isolated from different geographical locations from both Euprymna and Sepiola squid hosts and two monocentrid fishes, as well as some free-living isolates. All sequences were used to construct individual and combined phylogenetic trees to determine the relatedness amongst pil genes and to determine whether these loci can reveal whether host specificity via pili is a common character between phylogenetically distinct V. fischeri strains. In addition, we established the gene order in each of the strains tested to determine whether differences existed in the pil operons of free-living and symbiotic V. fischeri.
METHODS
Bacterial strains
Bacteria used in this study were isolated previously from a number of sepiolid squids and monocentrid fishes, as well as free-living strains from seawater (Table 1) (Nishiguchi & Nair, 2003). Bacteria were initially plated from a frozen glycerol stock onto Luria– Bertani high-salt (32 %) (LBS) agar and grown at their optimum temperature (28 uC). A single colony-forming unit (c.f.u.) was then used to inoculate 5 ml LBS, grown for 12 h overnight at the optimum growth temperature and used subsequently for DNA isolation. Chromosomal DNA was extracted using the DNeasy kit (Qiagen) as described by the manufacturer. DNA was determined to be of PCR quality by electrophoresis on 1.0% agarose gels.
Table 1. Bacterial isolates used in this study.
Accession numbers respectively represent 16S rRNA gene and pilB, pilC and pilD sequences unless indicated. –, Not sequenced.
| Strain | Host | Source | GenBank accession numbers |
|---|---|---|---|
| V. fischeri ATCC 7744 | Free living | Not known | AY292938, –, –, DQ207634 |
| V. fischeri ESP915 | Euprymna scolopes | USA (Paiko, HI) | AY292920, DQ093340 (B, C, D) |
| V. fischeri ES114 | Euprymna scolopes | USA (Kaneohe Bay, HI) | AY292919, DQ093345 (B, C, D) |
| V. fischeri EM17 | Euprymna morsei | Japan (Tokyo Bay) | AY292922, DQ093341 (B, C, D) |
| V. fischeri WH1 | Free living | USA (Woods Hole, MA) | AY292930, DQ093344 (B, C, D) |
| V. fischeri CG101 | Cleidopus gloriamaris | Australia (Townsville) | AY292939, DQ207619, DQ207627, DQ207636 |
| V. fischeri ET101 | Euprymna tasmanica | Australia (Crib Point) | AY292923, DQ093342 (B, C, D) |
| V. fischeri ET301 | Euprymna tasmanica | Australia (Botany Bay) | –, DQ207620, –, – |
| V. fischeri ET401 | Euprymna tasmanica | Australia (Magnetic Island) | AY292943, DQ207621, DQ207628, DQ207637 |
| V. fischeri ETJB5 | Euprymna tasmanica | Australia (Jervis Bay) | –, DQ207622, DQ207629, DQ207638 |
| V. fischeri SR5 | Sepiola robusta | France (Banyuls sur mer) | AY292926, DQ093343 (B, C, D) |
| V. fischeri SI1D | Sepiola intermedia | France (Banyuls sur mer) | AY292948, DQ207625, DQ207632, DQ207635 |
| V. fischeri SL518 | Sepiola ligulata | France (Banyuls sur mer) | AY292950, DQ207626, DQ207633, DQ207641 |
| V. fischeri MJ101 | Monocentrus japonicus | Japan | AY292946, DQ207624, DQ207631, DQ207640 |
| V. fischeri MDR7 | Free living | USA (Marina del Rey, CA) | AY292945, DQ207623, DQ207630, DQ207639 |
| Photobacterium leiognathi RM1 | Rondeletiola minor | France (Banyuls sur mer) | AY292947, –, –, – |
PCR amplifications
Individual loci of pilin genes were amplified by PCR in 25 µl reaction volumes. Primers were designed on the basis of the V. fischeri ES114 genome sequence (Ruby et al., 2005) (http://www.ncbi.nlm.nih.gov/sites/entrez?Db=genomeprj&Cmd=Retrieve&list_uids=12986). Primers used in these experiments are listed in Table 2. All PCRs contained 2.5 mM MgCl2, 1× reaction buffer (10 mM Tris/HCl, pH 9.0, 50 mM KCl, 0.1% Triton X-100), 2.5 mM each dNTP (Promega), 0.2 µM of each primer and 0.2 U Taq polymerase (Promega) or AmpliTaq (Applied Biosystems). PCR conditions are described in Table 3. All reactions included a hot start for 2 min at 94 uC and a final termination for 7 min at 72 °C using a DNA Engine Dyad (MJ Research). Amplified products were visualized via gel electrophoresis on a 1% agarose gel stained with ethidium bromide.
Table 2. Primers used in this study for amplification of pil loci.
Primers designed in this study were based on the V. fischeri ES114 genome sequence, as described in the text.
| Primer | Sequence (5′–3′) | Tm (°C) | Reference |
|---|---|---|---|
| ESpilA47 | GCAGAGCTCATATTAAAGCTCCTTCGTG | 64.6 | Stabb & Ruby (2003) |
| ESpilA48 | CGTCGTATTGAGGCGATGCGTGAG | 70.7 | Stabb & Ruby (2003) |
| PilC2 | CCAACTTGAGGGCTTGCACAATGG | 66.3 | This study |
| PilC3 | GTAACGAGTGTACGGGCGGCTATT | 66.3 | This study |
| JBSpilB F | GCACACCAACCTCGCCTC | 59.3 | This study |
| JBSpilB R | GAGCACGGTTTAGTTCTTG | 51.9 | This study |
| JBSpilC F | GCTTTCAATGGCGTGGC | 55.6 | This study |
| JBSpilC R | GCCAATAACACTCATAAG | 45.7 | This study |
| JBSpilD F | GATTATTACCCTTGGCTT | 41.7 | This study |
| JBSpilD R | GTGGCAAACCTAGAATG | 48.0 | This study |
| pilC 3′1 | CCGCCGGTAATCTTTACTATCAAGAAGCCA | 63.7 | This study |
| 5′1pilC | TGAACAACWGGAACACCTGAGGCGAGC | 64.1 | This study |
| pilB1-175 | GAACTCCACTAGTTGCACTGAGTTAAAACT | 60.7 | This study |
| pilD1-3860 | GCTATTGCTTGGGTTTGCCTATTATGG | 65.1 | This study |
| AP1 | GTAATACGACTCACTATAGGC | 59.0 | BD Biosciences |
Table 3.
PCR conditions used in this study
| Gene target | Cycles (n) | Temperature (°C) | ||
|---|---|---|---|---|
| Denaturing | Annealing | Extension | ||
| pilA | 35 | 94 | 58 | 72 |
| pilB | 30 | 94 | 49 | 72 |
| pilC (primers PilC2 and PilC3) | 30 | 94 | 53 | 72 |
| pilC (other primers) | 30 | 94 | 43 | 72 |
| pilD | 30 | 94 | 43 | 72 |
Southern blotting
Southern blotting was used to test whether strains of V. fischeri that had unsuccessful PCR amplification of pil genes still possessed homologous regions of the pil operon. Genomic DNA was digested with HindIII, run on a 1% agarose gel and transferred to nylon blotting paper (Millipore) by a standard method (Sambrook et al., 1989). Each blot was probed with ESP915pilA (designed from the V. fischeri ES114 genome) and labelled with 32P. Reactions contained 10 pmol oligonucleotide (5.0 µl), 2.0 µl kinase buffer (50 mM Tris/HCl, 10 mM MgCl2, 0.1 mM EDTA, 5 mM DTT, 0.1 mM spermidine), 2.0 µl sterile distilled water, 0.5 U polynucleotide kinase (1.0 µl) and 10 µl [γ-32P]ATP (100 µCi; Amersham Quebec). Labelling reactions were incubated at 37 °C for 30 min. Membranes were hybridized overnight individually with each probe at the specific activity of 5 × 105 µCi ml−1 at 48 °C. The hybridization solution was discarded and the membranes were washed twice for 15 min each with washing buffer at 48 °C. Membranes were then exposed to X-ray film for 3 h to determine the presence of the pil operon.
Cloning and sequencing
Reactions that produced the desired PCR product were purified with the Geneclean II kit (Bio 101) and sequenced using Applied Biosystems Big Dye (version 3.1). PCR products that proved difficult to sequence directly were cloned into pGEM-T Easy vector (Promega) using the manufacturer’s protocol. Plasmid DNA from positive clones was extracted using the Qiagen miniprep kit and sequenced. Excess fluorescently labelled dNTPs were removed via spin columns or plates (Edge Biosystems) and DNA was subsequently sequenced using an Applied Biosystems 3100 automated sequencer. Accession numbers for all sequences generated are listed in Table 1.
Gene walking to determine gene order
New primers corresponding to the 5′ and 3′ ends of each pil locus were designed using the derived sequences to link and determine the order of individual pil loci. Using the Universal Genome Walker kit (BD Biosciences), chromosomal DNA was digested separately with four restriction enzymes, DraI, EcoRV, PvuII and StuI. The reactions were purified with 100% saturated phenol (pH 8.0), with the upper aqueous layer re-extracted with chloroform. DNA was precipitated with 2 vols cold 95% ethanol, 20 µg glycogen and 0.1 vols 3 M sodium acetate. The mixture was centrifuged at 12 000 g for 10 min and dissolved in TE (10 mM Tris/HCl, 0.1 mM EDTA, pH 7.5).
Ligation reactions included 4 µl digested DNA, 25 µM GenomeWalker adaptor, 1.6 µl ligation buffer and 0.5 µl (3 U) T4 DNA ligase. Reactions were performed overnight at 16 °C in a DNA Engine Dyad (MJ Research). Samples were diluted 1 : 100 with sterile distilled water and used for PCRs. PCR was performed using Herculase Taq DNA polymerase (Stratagene), AP1 primer and individual pilC primers (Table 2). The PCR was completed in 25 µl reaction volumes containing 2.5 µl 10× reaction buffer, 0.2 µl dNTPs (25 mM each), 0.5 µg each primer and 1.25 U Herculase Taq polymerase. The following conditions were followed: seven cycles at 94 °C for 25 s and the primer Tm for 3 min; 32 cycles of 94 °C for 25 s and the primer Tm minus 3 °C for 3 min and a final extension at 61 °C for 7 min. PCR products were purified using Geneclean II and sequenced directly (ABI Big Dye v. 3.1) or cloned into pGEM-T Easy vector as described above. Fluorescently labelled products were purified using Edge DTR columns (Edge Biosystems) and analysed on an ABI 3100 automated sequencer.
Sequence alignment and phylogenetic analyses
Double-stranded DNA sequences were assembled and edited using the program Sequencher version 4.0 (Gene Codes). Sequences were translated to determine the correct reading frame and promoter region to establish whether the genes were in one continuous operon. Comparison between strains was essential, since the arrangement of pil genes was not universal among all V. fischeri strains tested, as well as the presence of all loci. Each sequence was compared with the NCBI Blastx search engine to determine similarity to other available pil sequences.
Molecular data were analysed with the computer program POY (Wheeler et al., 2002) using the direct optimization method, with parsimony as the optimality criterion (Wheeler, 1996). This method assesses the number of DNA sequence transformations (evolutionary events) required by a phylogenetic topology directly, without the use of multiple sequence alignment. This is accomplished by generalization of existing character optimization procedures, including insertion and deletion events (indels) in addition to base substitutions. This method treats indels as processes, as opposed to the patterns implied by multiple sequence alignment (Wheeler, 1995).
Nodal support was calculated in POY using Farris’s parsimony jackknifing procedure (Farris et al., 1996) for 100 replicates (Using the commands –jackboot –replicates 100). Tree searches were conducted in parallel at Harvard University on a 19 dual-processor cluster using pvm (parallel virtual machine). Commands for load balancing of spawned jobs were used to optimize parallelization procedures (–parallel –dpm –jobspernode 2). Trees were built via a random addition sequence procedure (10 replicates) followed by a combination of branch-swapping steps [SPR (subtree pruning and regrafting) and TBR (tree bisection and reconnection)] and tree fusing (Goloboff, 1999) in order to improve tree length minimization further. Discrepancies between heuristic and actual tree length calculations were addressed by adjusting slop values (–slop5 – checkslop10). Phylogenetic trees were obtained using parsimony with a gap/ts/tv cost of equal weighting. The final tree was drawn with TreeView (Win32) and consensus trees were analysed in PAUP version 4.02b (Swofford, 2002). Nodal support for all jackknife calculations was performed in POY by the procedure described in Nishiguchi & Nair (2003). Individual gene trees for each of the pil loci were produced, as well as total molecular trees combining all pil loci and all pil loci with the 16S rRNA gene.
RESULTS AND DISCUSSION
Analysis of pil gene order and content
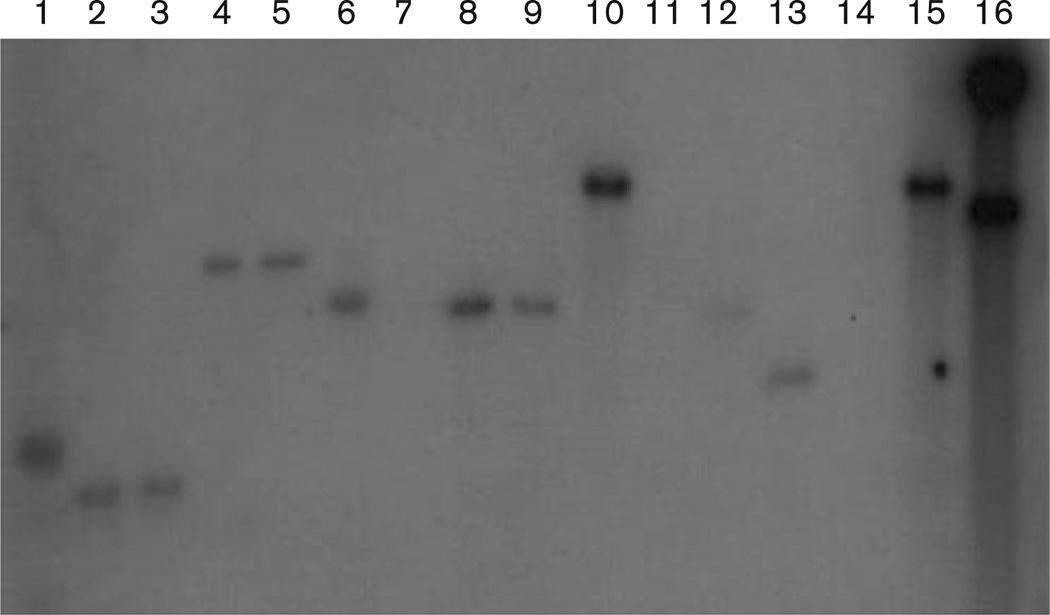
The amplified pilA gene from V. fischeri ESP915 was used as a probe (designed in this study) and hybridized to 2.5 kb fragments from V. fischeri strains EM17, ES114, ET101, SA1G and SR5 using Southern blot verification. This demonstrated the presence of a homologous pilA gene in those strains. In addition, a 4.5–5.0 kb band was observed in V. fischeri strains EB12 and WH1 and Vibrio logei strain SR18-1 (Fig. 1). This is expected, since previous evidence had demonstrated that the V. fischeri genome contains ten individual clusters of pil genes, of which two are pilA homologues (Ruby et al., 2005). Although similarities existed among the symbiotic strains of V. fischeri probed, no hybridization occurred with DNA from either Vibrio harveyi ATCC 14126T or Vibrio cholerae RGELP40. This may be due to the fact that both V. cholerae and V. harveyi are pathogenic bacteria, and may have evolved a different pilin structure necessary for the infection of their respective host tissues. In a phylogenetic context, V. cholerae and V. harveyi are not closely related to one another or to V. fischeri, which again provides evidence that colonization and infection mechanisms have evolved multiple times within the Vibrionaceae (Nishiguchi & Nair, 2003; Thompson et al., 2004a).
Fig. 1.
Southern blot of HindIII digests probed with the pilA gene from V. fischeri ESP915. Lanes: 1, 1 kb ladder; 2, V. fischeri EB12; 3, V. fischeri EM17; 4, V. fischeri ES114; 5, V. fischeri ET101; 6, V. fischeri SA1G; 7, V. logei SR18-1; 8, V. fischeri SR5; 9, V. fischeri ESP915; 10, V. harveyi ATCC 14126T; 11, Photobacterium leiognathi RM1; 12, V. fischeri WH1; 13, V. parahaemolyticus ATCC 17802T; 14, V. logei ATCC 35077; 15, E. coli O157 : H7-84/24; 16, V. cholerae RGELP40.
Availability of the V. fischeri ES114 genome sequence also enabled the design of primers for the pilB, pilC and pilD loci. Targeted genes were amplified as well as an additional 350 bp from V. fischeri strains ET101, ESP915 and WH1. V. fischeri EB12 produced an additional 650 bp fragment, while V. fischeri strains ET101, SA1G, SA1J and SI1E produced an additional 550 bp band with the JBSpilB primers (Table 2). All V. fischeri strains except for ATCC 7744, a free-living isolate, were successfully amplified with the JBSpilC primers. Vibrio parahaemolyticus ATCC 17802T, Photobacterium phosphoreum PP21 and V. fischeri ATCC 7744 yielded a smaller 850 bp fragment, with sequence similarity to other pilB sequences. All of these vibrios as well as V. fischeri ATCC 7744 cannot colonize squid light organs (M. K. Nishiguchi, unpublished) and may therefore be missing an important functional group of the pil gene cluster that enables symbiotic bacteria to colonize animal tissues successfully.
When comparing all V. fischeri DNA sequences from this study, it was noted that all sequences were A+T rich (61 mol%). For the pilA locus, a 740 bp fragment was amplified and sequenced from all bacteria. Blastx demonstrated that the initial 502 bp was related to pilA and the remaining 309 bp was homologous to the polar flagellar sequence of other vibrios, with the only exception being V. harveyi. clustal w alignment demonstrated that pilA was 86–95% conserved in nucleotide sequence among all strains examined in this study. A multiple alignment of the in-frame amino acid sequences exhibited the same conservation. Results for the pilB comparisons gave ≥93% similarity. This was not surprising, since previous evidence has demonstrated that pilA can be replaced by corresponding genes from non-transformable species (Graupner et al., 2000). The pilA locus is a pre-pilin gene that is flanked by other pil-like genes in numerous Gram-negative bacteria but, in the V. fischeri strains examined, no other pil-like genes were found in the vicinity (Stabb & Ruby, 2003). From our genome-walking experiments, only proteins whose functions are unknown (as determined for the V. fischeri ES114 genome; Ruby et al., 2005) were within the 1000 bp region examined. Previous studies in other Gram-negative bacteria have indicated that the pilA and pilD genes are 100% conserved, with variation observed most widely at the pilC locus (Fullner & Mekalanos, 1999). We found that the pilC genes sequenced in this study were 94–97% similar, with the exception of V. fischeri WH1 and V. fischeri MDR7, both of which are freeliving isolates (100% sequence similarity to each other at the pilC locus, but less than 90% similarity to symbiotic isolates). pilC has been one of the pil loci identified as critical for adhesin in the binding of Neisseria gonorrhoeae to human epithelial cells (Rudel et al., 1995b). Likewise, a pilC gene homologue has also been observed in Pseudomonas aeruginosa and has been linked to virulence in this species (Rudel et al., 1995a; Alm et al., 1996), suggesting that the pilC gene is important for the infectivity of these pathogenic bacteria. Interestingly, neither of our free-living strains (WH1 and MDR7) can colonize or persist inside squid light organs, and the 10% sequence difference observed at this locus may account for a loss of symbiotic competence. Future experiments to complement free-living V. fischeri strains such as WH1 and MDR7 with pil genes may help to determine whether improved colonization can be achieved with this operon alone, or whether other factors may be responsible for colonization and persistence in squid light organs.
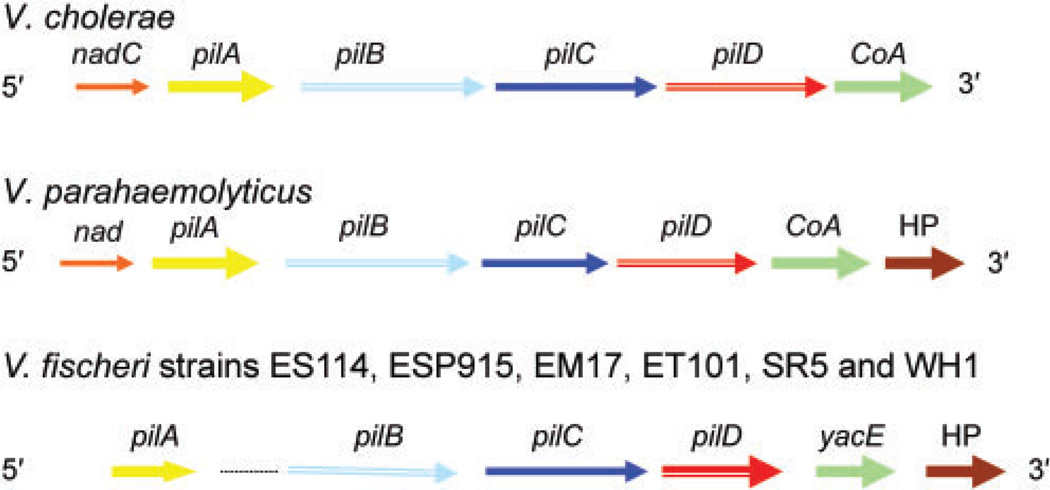
The order of pil genes in the V. fischeri strains examined also differed from pathogenic V. cholerae and V. parahaemolyticus. pilA was determined to be monocistronic in V. fischeri and was separated from the other pil loci, which were present downstream and not in one continuous reading frame (Fig. 2). This is different from pilA found in both V. cholerae and V. parahaemolyticus. pilB (1674 bp) and pilC (1239 bp) were both translated in the second reading frame, while pilD (933 bp) was translated in the first reading frame. There were 19 bases between pilB and pilC (the pilin assembly/biogenesis protein), 59 bases between pilC and pilD (the pilin peptidase), a 24 base overlap of the dephospho-CoA kinase (CoA, or yacE in V. cholerae) and 6 bases between yacE and a conserved hypothetical protein (HP) when compared to the V. fischeri ES114 genome. Using PsortB (http://psort.nibb.ac.jp/form.html), the following predictions were made for the V. fischeri strains examined in this study: (i) PilB has the conformation of a cytoplasmic protein, (ii) PilC is similar to an internal membrane protein that contains four internal helices and (iii) PilD has the conformation of an inner-membrane protein (Pepe & Lory, 1998). Additionally, a fragment which proved to be homologous to the membrane-bound phosphotransferase system (pts), which phosphorylates the manXYZ operon (Erni & Zanolari, 1985), was found to follow the hypothetical protein (not shown).
Fig. 2.
Gene order of each of the pil operons from five symbiotic (ES114, ES915, EM17, ET101 and SR5) and one free-living (WH1) strain of V. fischeri compared with V. cholerae RGELP40 and V. parahaemolyticus ATCC 14126T. Sequences of ~5 kb were utilized to construct the gene maps for all strains.
Phylogeny of symbiotic vibrios using the pil operon
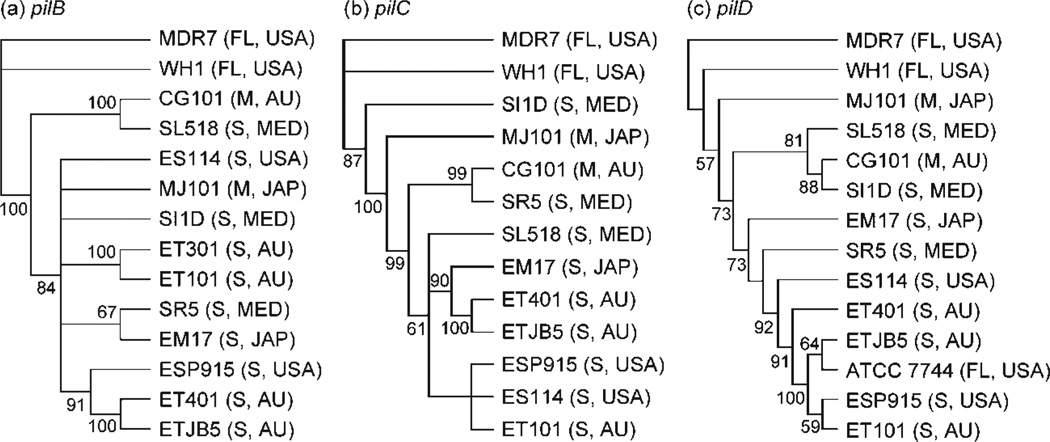
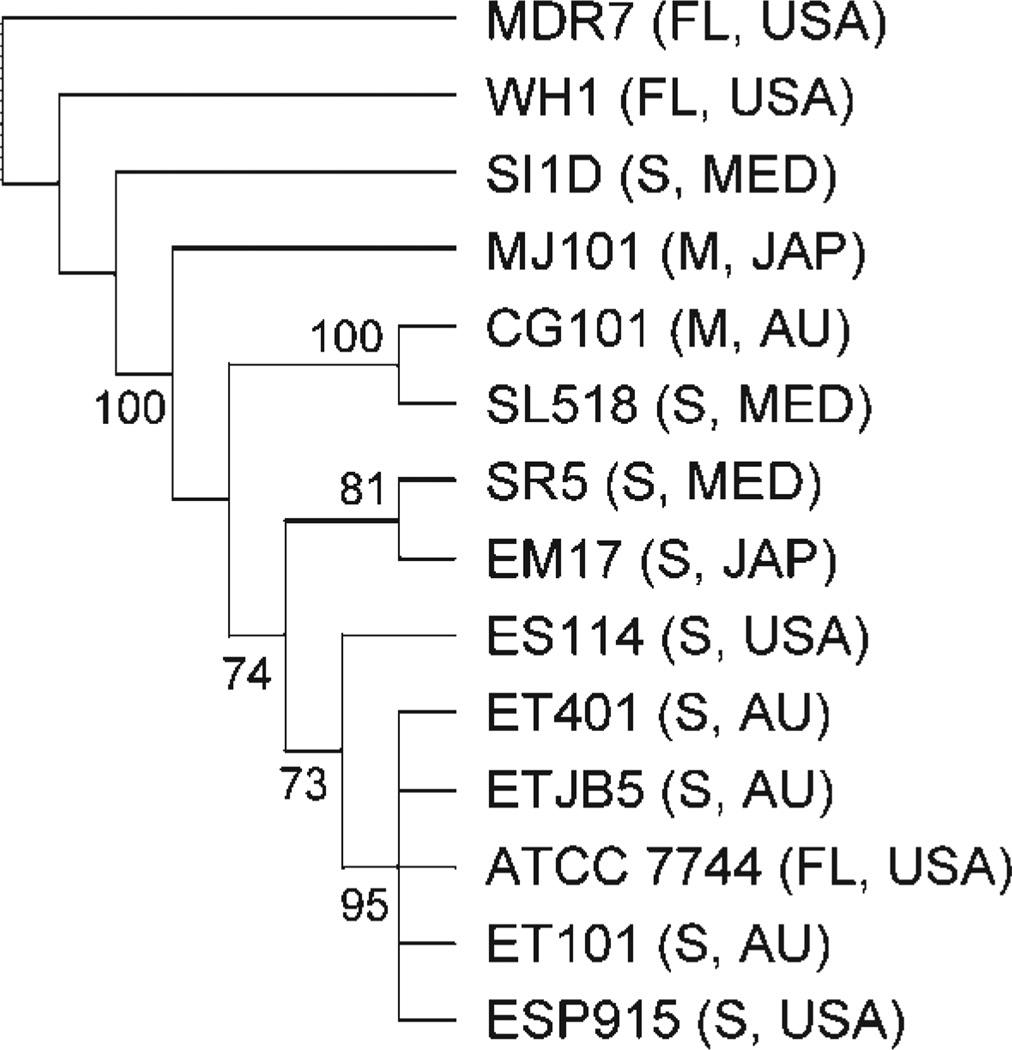
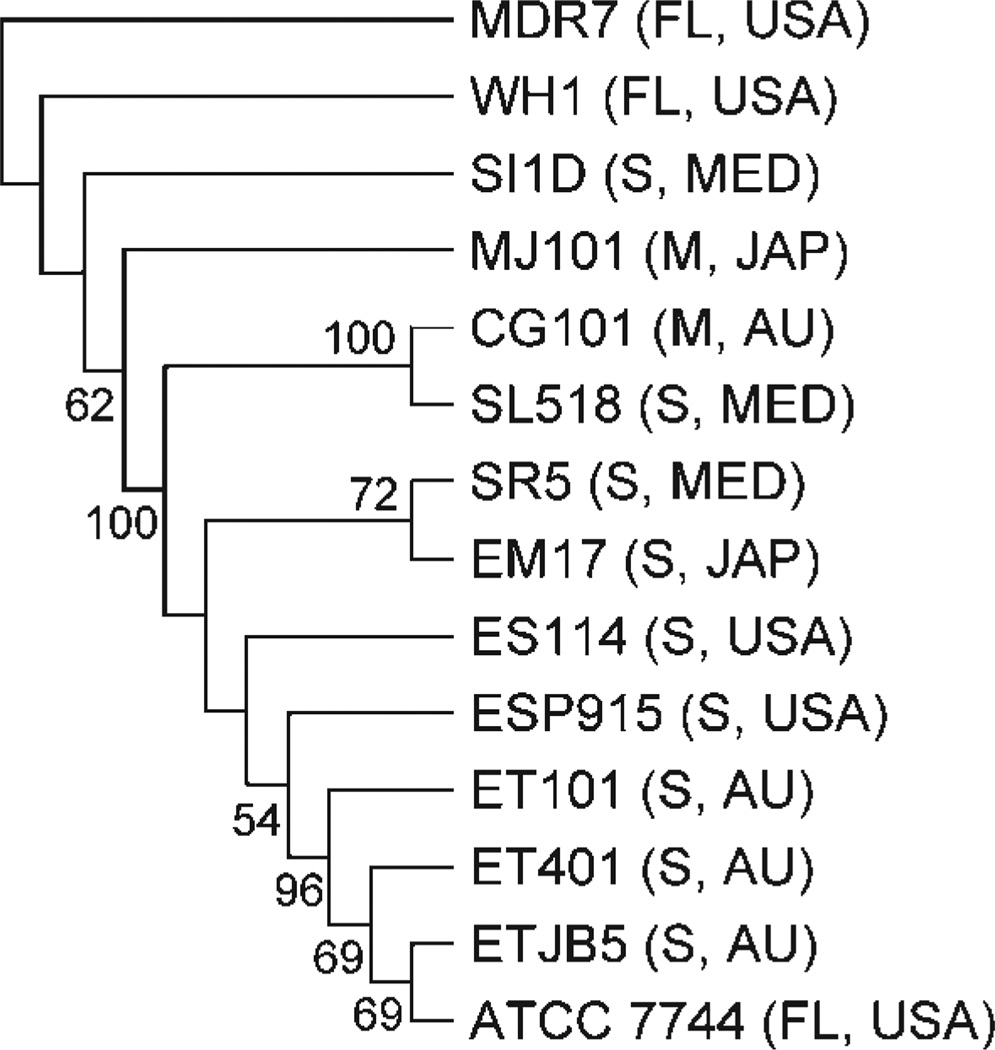
All individual and total molecular gene trees were outgrouped with V. fischeri WH1 and MDR7, both of which are free-living isolates. We did not use V. fischeri ATCC 7744 as a free-living outgroup, since earlier studies suggested that this strain was divergent from the other V. fischeri strains used in this study (Nishiguchi & Nair, 2003). Phylogenetic trees based on individual pil loci produced phylogenies that were similar to each other, with most Indo-west Pacific vibrios (‘E’ strains) grouping together (Fig. 3). This was expected from our sequence analysis, since individual sequences for each locus were highly conserved with little variation. The two symbiotic fish strains (MJ101 and CG101) did not group together in all three individual gene analyses, or in the combined molecular analysis of all pil loci (Fig. 4). The three Mediterranean strains (‘S’ strains) analysed in our study were also included in clades containing fish and squid ‘E’ strains (Fig. 3). Thus, no geographical or host separation was made evident by any single gene phylogeny or the combined phylogenetic analysis of the entire pil gene operon (Fig. 4). The combined pil gene analysis (pilB, pilC and pilD) did group all ET and ES strains into a single clade, but this clade also included V. fischeri SR5 (Mediterranean) and ATCC 7744 (Fig. 4). A combined analysis of pilB, pilC, pilD and the 16S rRNA locus also indicated that all ET and ES strains were grouped within one clade (again including SR5 and ATCC 7744; Fig. 5).
Fig. 3.
Gene trees for pilB (a), pilC (b) and pilD (c) for equal-weighted parameter sets (indels/is/tv) that minimized overall incongruence using POY with parsimony as the criterion. Jacknife values >50% are shown at nodes. Outgroups are V. fischeri strains MDR7 and WH1 (free-living). FL, Free-living; S, sepiolid symbiont; M, monocentrid symbiont; USA, United States isolate; AU, Australian isolate; MED, Mediterranean Sea isolate; JAP, Japanese isolate. See Table 1 for sequence accession numbers.
Fig. 4.
Total combined pil loci (B, C and D) tree for equal-weighted parameter set (indels/ts/tv) that minimized overall incongruence of the three genes using POY with parsimony as the criterion. See legend to Fig. 3 for further details.
Fig. 5.
Total molecular tree combining all pil loci (B, C and D) and the 16S rRNA locus for equal-weighted parameter set (indels/ts/tv) that minimized the overall incongruence of the four genes using POY with parsimony as the criterion. See legend to Fig. 3 for further details.
Both combined tree analyses verify previous work using haplotype comparisons and nested clade analysis that demonstrated that ET and ES strains are closely related to one another and occupy populations of Euprymna that span a large geographical range (Hawaii, Australia and Thailand; Jones et al., 2006). Vibrio strains did not cluster together based on geographical location or host specificity, as hypothesized previously based on competition experiments (Nishiguchi et al., 1998; Nishiguchi, 2002). However, these earlier studies were based on a single clone isolated from an individual squid host and were not representative of the large genetic breadth exhibited by Vibrio (Thompson et al., 2004b; Thompson & Swings, 2006). Likewise, with more Vibrio genomes becoming available, there is growing evidence that horizontal gene transfer events and the number of increased mutation events (particularly those Vibrio species that are subject to host control, such as V. fischeri) are responsible for the rapid radiation of this dynamic group of bacteria. Recent work has demonstrated that vibrios have the capability to expand their environmental niches (both free-living and symbiotic; Jones et al., 2006), which may explain why so many symbiotic V. fischeri strains have the capability to infect other species of host squid (Nishiguchi, 2002). In addition, recent work has demonstrated that V. harveyi is also capable of infecting and colonizing the light organs of loliginid squids (Guerrero-Ferreira & Nishiguchi, 2007), which exemplifies how Vibrio bacteria are adept at expanding into new ecological niches. The fact that V. fischeri and V. logei had previously been identified as the only two species of light-organ-inhabiting bacteria illustrates how little we know about the vast number of ecological niches that are available to vibrios and whether they are more susceptible to environmental selection or horizontal gene transfer events that may lead to incongruence in phylogenetic topologies (Thompson et al., 2004b). Although this study was focused mainly on determining whether pilin gene sequences could distinguish between closely related V. fischeri strains, further studies need to be completed that will determine whether changes in any of the pil genes influence colonization specificity observed among closely related Vibrio symbionts. This would include constructing mutations within the pil genes to determine how deletions will affect adhesion and colonization specificity. Also, in vivo adherence assays of mutant V. fischeri strains to light organ epithelia cells would determine whether a change in colonization efficiency between wild-type and mutant vibrios is a main factor for driving recognition among closely related host–symbiont pairs. Whether variability between these genes is caused by rapid evolution, host selection or other gene transfer events remains to be determined and will provide yet another window into the evolution of this interesting group of gammaproteobacteria.
ACKNOWLEDGEMENTS
The authors would like to thank E. Stabb for advice on pilA primers in this project. R. DeVinney provided our E. coli strains used as controls in this project. This research was supported by grant SO6-GM0813626 from the National Institutes of Health and NSF SBE-0123690 and DEB-0316516 to M. K. N. We would like to thank the support of undergraduates Kitty Brown, Rebecca Lopez, Jonathan Clyde and Charity Watching, who were supported by either NIH-MARC (GM0766724) or NIH-BRIDGES (GM48998).
Footnotes
The GenBank/EMBL/DDBJ accession numbers for the sequences determined in this study are detailed in Table 1.
REFERENCES
- Alm RA, Hallinan JP, Watson AA, Mattick JS. Fimbrial biogenesis genes of Pseudomonas aeruginosa: pilW and pilX increase the similarity of type 4 fimbriae to the GSP protein-secretion systems and pilY1 encodes a gonococcal PilC homologue. Mol Microbiol. 1996;22:161–173. doi: 10.1111/j.1365-2958.1996.tb02665.x. [DOI] [PubMed] [Google Scholar]
- Darzins A, Russell MA. Molecular genetic analysis of type-4 pilus biogenesis and twitching motility using Pseudomonas aeruginosa as a model system – a review. Gene. 1997;192:109–115. doi: 10.1016/s0378-1119(97)00037-1. [DOI] [PubMed] [Google Scholar]
- Erni B, Zanolari B. The mannose-permease of the bacterial phosphotransferase system. Gene cloning and purification of the enzyme IIMan/IIIMan complex of Escherichia coli. J Biol Chem. 1985;260:15495–15503. [PubMed] [Google Scholar]
- Farris JS, Albert VA, Källersjö M, Lipscomb D, Kluge AG. Parsimony jackknifing outperforms neighbor-joining. Cladistics. 1996;12:99–124. doi: 10.1111/j.1096-0031.1996.tb00196.x. [DOI] [PubMed] [Google Scholar]
- Fullner KJ, Mekalanos JJ. Genetic characterization of a new type IV-A pilus gene cluster found in both classical and El Tor biotypes of Vibrio cholerae. Infect Immun. 1999;67:1393–1404. doi: 10.1128/iai.67.3.1393-1404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goloboff PA. Analyzing large data sets in reasonable times: solutions for composite optima. Cladistics. 1999;15:415–428. doi: 10.1111/j.1096-0031.1999.tb00278.x. [DOI] [PubMed] [Google Scholar]
- Graf J, Dunlap PV, Ruby EG. Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J Bacteriol. 1994;176:6986–6991. doi: 10.1128/jb.176.22.6986-6991.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graupner S, Frey V, Hashemi R, Lorenz MG, Brandes G, Wackernagel W. Type IV pilus genes pilA and pilC of Pseudomonas stutzeri are required for natural genetic transformation, and pilA can be replaced by corresponding genes from nontransformable species. J Bacteriol. 2000;182:2184–2190. doi: 10.1128/jb.182.8.2184-2190.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Ferreira RC, Nishiguchi MK. Biodiversity among luminescent symbionts from squid of the genera Uroteuthis, Loliolus and Euprymna (Mollusca: Cephalopoda) Cladistics. 2007;23:497–506. doi: 10.1111/j.1096-0031.2007.00155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B, Nishiguchi M. Counterillumination in the Hawaiian bobtail squid, Euprymna scolopes Berry (Mollusca: Cephalopoda) Mar Biol. 2004;144:1151–1155. [Google Scholar]
- Jones BW, Huttenberg J, Lopez JE, Nishiguchi MK. Population structure between environmentally transmitted vibrios and bobtail squids using nested clade analysis. Mol Ecol. 2006;15:4317–4329. doi: 10.1111/j.1365-294X.2006.03073.x. [DOI] [PubMed] [Google Scholar]
- McFall-Ngai MJ. Animal-bacterial interactions in the early life history of marine invertebrates: the Euprymna scolopes/Vibrio fischeri symbiosis. Am Zool. 1994;34:554–561. [Google Scholar]
- McFall-Ngai MJ, Ruby EG. Sepiolids and vibrios: when first they meet. Bioscience. 1998;48:257–265. [Google Scholar]
- Millikan DS, Ruby EG. Vibrio fischeri flagellin A is essential for normal motility and for symbiotic competence during initial squid light organ colonization. J Bacteriol. 2004;186:4315–4325. doi: 10.1128/JB.186.13.4315-4325.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair VS. PhD dissertation. Las Cruces, NM, USA: New Mexico State University; 2006. The evolution of symbiosis in Vibrionaceae and factors influencing host colonization in a squid-Vibrio mutualism. [Google Scholar]
- Nishiguchi MK. Temperature affects species distribution in symbiotic populations of Vibrio spp. Appl Environ Microbiol. 2000;66:3550–3555. doi: 10.1128/aem.66.8.3550-3555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiguchi MK. Host-symbiont recognition in the environmentally transmitted sepiolid squid-Vibrio mutualism. Microb Ecol. 2002;44:10–18. doi: 10.1007/BF03036870. [DOI] [PubMed] [Google Scholar]
- Nishiguchi MK, Nair VS. Evolution of symbiosis in the Vibrionaceae: a combined approach using molecules and physiology. Int J Syst Evol Microbiol. 2003;53:2019–2026. doi: 10.1099/ijs.0.02792-0. [DOI] [PubMed] [Google Scholar]
- Nishiguchi MK, Ruby EG, McFall-Ngai MJ. Competitive dominance among strains of luminous bacteria provides an unusual form of evidence for parallel evolution in sepiolid squid-Vibrio symbioses. Appl Environ Microbiol. 1998;64:3209–3213. doi: 10.1128/aem.64.9.3209-3213.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn D. Bacterial type II protein export and pilus biogenesis: more than just homologies? Trends Cell Biol. 1999;9:402–408. doi: 10.1016/s0962-8924(99)01634-7. [DOI] [PubMed] [Google Scholar]
- Nyholm SV, McFall-Ngai MJ. The winnowing: establishing the squid-Vibrio symbiosis. Nat Rev Microbiol. 2004;2:632–642. doi: 10.1038/nrmicro957. [DOI] [PubMed] [Google Scholar]
- Pepe JC, Lory S. Amino acid substitutions in PilD, a bifunctional enzyme of Pseudomonas aeruginosa. Effect on leader peptidase and N-methyltransferase activities in vitro and in vivo. J Biol Chem. 1998;273:19120–19129. doi: 10.1074/jbc.273.30.19120. [DOI] [PubMed] [Google Scholar]
- Ruby EG, Urbanowski M, Campbell J, Dunn A, Faini M, Gunsalus R, Lostroh P, Lupp C, McCann J, et al. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc Natl Acad Sci U S A. 2005;102:3004–3009. doi: 10.1073/pnas.0409900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudel T, Boxberger HJ, Meyer TF. Pilus biogenesis and epithelial cell adherence of Neisseria gonorrhoeae pilC double knock-out mutants. Mol Microbiol. 1995a;17:1057–1071. doi: 10.1111/j.1365-2958.1995.mmi_17061057.x. [DOI] [PubMed] [Google Scholar]
- Rudel T, Facius D, Barten R, Scheuerpflug I, Nonnenmacher E, Meyer TF. Role of pili and the phase-variable PilC protein in natural competence for transformation of Neisseria gonorrhoeae. Proc Natl Acad Sci U S A. 1995b;92:7986–7990. doi: 10.1073/pnas.92.17.7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: a Laboratory Manual. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- Sandkvist M. Biology of type II secretion. Mol Microbiol. 2001;40:271–283. doi: 10.1046/j.1365-2958.2001.02403.x. [DOI] [PubMed] [Google Scholar]
- Soto GE, Hultgren SJ. Bacterial adhesins: common themes and variations in architecture and assembly. J Bacteriol. 1999;181:1059–1071. doi: 10.1128/jb.181.4.1059-1071.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabb EV, Ruby EG. Contribution of pilA to competitive colonization of the squid Euprymna scolopes by Vibrio fischeri. Appl Environ Microbiol. 2003;69:820–826. doi: 10.1128/AEM.69.2.820-826.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. paup*: Phylogenetic analysis using parsimony (and other methods), version 4. Sunderland, MA: Sinauer Associates; 2002. [Google Scholar]
- Thompson FL, Swings J. Taxonomy of the vibrios. In: Thompson FL, Austin B, Swings J, editors. The Biology of Vibrios. Washington, DC: American Society for Microbiology; 2006. pp. 29–43. [Google Scholar]
- Thompson CC, Thompson FL, Vandemeulebroecke K, Hoste B, Dawyndt P, Swings J. Use of recA as an alternative phylogenetic marker in the family Vibrionaceae. Int J Syst Evol Microbiol. 2004a;54:919–924. doi: 10.1099/ijs.0.02963-0. [DOI] [PubMed] [Google Scholar]
- Thompson FL, Iida T, Swings J. Biodiversity of vibrios. Microbiol Mol Biol Rev. 2004b;68:403–431. doi: 10.1128/MMBR.68.3.403-431.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar MT, Hirschberg RL, Schaefer MR. Role of the Eikenella corrodens pilA locus in pilus function and phase variation. J Bacteriol. 2001;183:55–62. doi: 10.1128/JB.183.1.55-62.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visick KL, Ruby EG. Vibrio fischeri and its host: it takes two to tango. Curr Opin Microbiol. 2006;9:632–638. doi: 10.1016/j.mib.2006.10.001. [DOI] [PubMed] [Google Scholar]
- Wheeler WC. Sequence alignment, parameter sensitivity, and the phylogenetic analysis of molecular data. Syst Biol. 1995;44:321–331. [Google Scholar]
- Wheeler W. Optimization alignment: the end of multiple sequence alignment in phylogenetics? Cladistics. 1996;12:1–9. [Google Scholar]
- Wheeler WC, Gladstein DS, DeLaet J. poy: the optimization of alignment characters. New York: American Museum of Natural History; 2002. [Google Scholar]
- Winther-Larsen HC, Hegge FT, Wolfgang M, Hayes SF, van Putten JP, Koomey M. Neisseria gonorrhoeae PilV, a type IV pilus-associated protein essential to human epithelial cell adherence. Proc Natl Acad Sci U S A. 2001;98:15276–15281. doi: 10.1073/pnas.261574998. [DOI] [PMC free article] [PubMed] [Google Scholar]