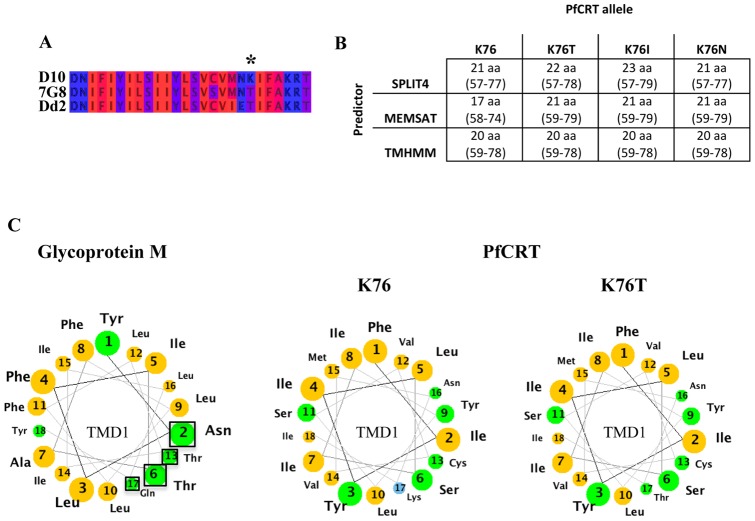
Figure 2. Bioinformatic analysis of the K76T mutation in PfCRT.
(A) Multiple sequence alignment (by Jalview [38]) of the first TMD of CQ-sensitive D10 and CQ-resistant 7G8 and Dd2 parasites [22]. The amino acids are represented according to their hydrophobicity (hydrophilic residue (blue), hydrophobic residue (red), intermediate stages (dark red, violet)). The mutation at position 76 represents a replacement of a positively charged, hydrophilic residue (lysine, K) with an uncharged, hydrophobic (threonine, T) residue. (B) Predicted length of the first TMD of sensitive and various resistant alleles of PfCRT using different bioinformatic predictors. Single point mutations at position 76 [22], [63] were analysed using three different TM prediction applications, i.e. TMHMM2 [39], Memsat [40] and Split4 [41]. Following the TM prediction the predicted length and position of the first TMD of PfCRT were extracted and analysed. In contrast to TMHMM2, the predictors Memsat and Split predict a change in both length and position due to a single point mutation at position 76. (C) Helical wheel projection of the first TMD of Glycoprotein M (left) and PfCRT (right). Yellow residues are nonpolar; green residues are polar, uncharged; blue residues are basic. The first TMD of the infectious bronchitis virus Glycoprotein M consists of a polar face (black square around the involved residues) within its first TMD that becomes obvious when represented in a helical wheel projection (modified from [42]). Point mutations that modified the polar face of the TMD resulted in a mistargeting of the viral Glycoprotein M [42]. Helical wheel projection of the first TMD of PfCRT reveals a change in the polar face as a result of mutations at position 76 (modified from [62]).