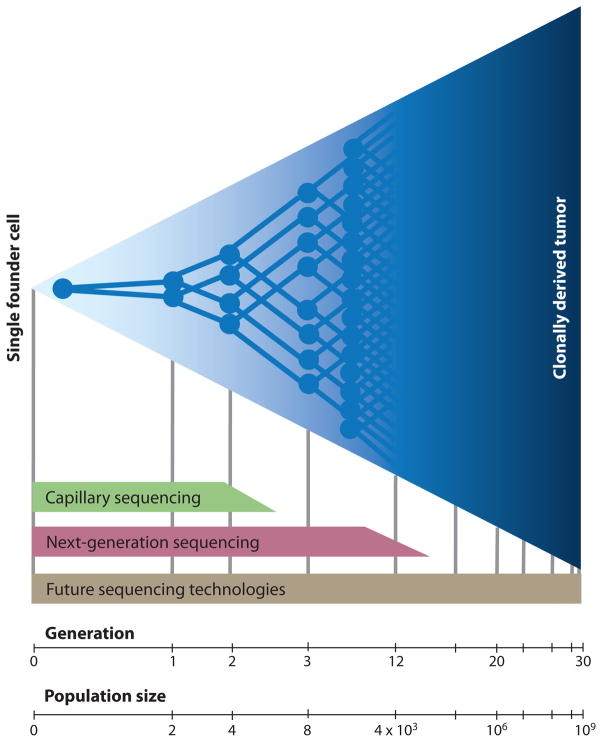
Figure 5. Limit of subclonal detection.
Depicted is the clonal expansion of a single cell into a population of one billion cells. In a hypothetical scenario where no cell death occurs, this requires approximately 30 generations of division. Current capillary methods of DNA sequencing only detect mutations that have clonally expanded to represent 25% or more of a tumor. Only mutations that are present in the founding cell or that arise within the first two generations of division can be identified. Deep sequencing on current “next-generation” sequencing platforms are reported to detect subclonal mutations down to a frequency of 1/5000 (119) (thus, those occurring within the first twelve generations after founding). Sensitivity is limited by the error rate of PCR amplification steps and that of the sequencing chemistry itself. Future technologies may eventually enable ultra-accurate, high-throughput detection of mutations that arise during any stage of clonal expansion.