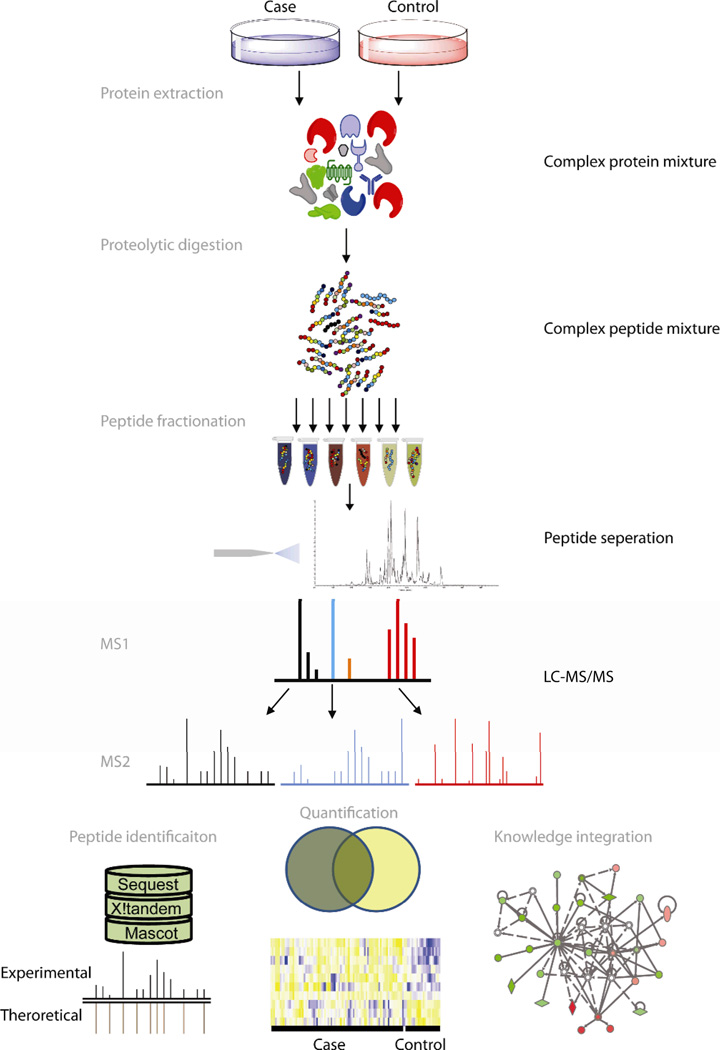
Figure 1.
Overview of bottom-up proteomics. In MS-based bottom-up ‘shotgun’ proteomics studies complex mixtures of proteins are isolated from the biological sample of interest and enzymatically or chemically cleaved into peptides. The peptide mixture is often fractionated and analyzed by tandem LC MS (MS/MS). Tandem LC-MS analysis involves the acquisition of a preliminary mass spectrum (MS1) of the intact (precursor) species, dissociation of selected ions of interest into smaller fragments, and then mass analysis of the fragments (MS2). Peptide identification of tandem MS spectra is often performed by matching mass measurements for intact peptides and MS/MS fragment ions to theoretical sequences derived from genome sequence information. Quantification and visualization tools facilitate interpretation of data.