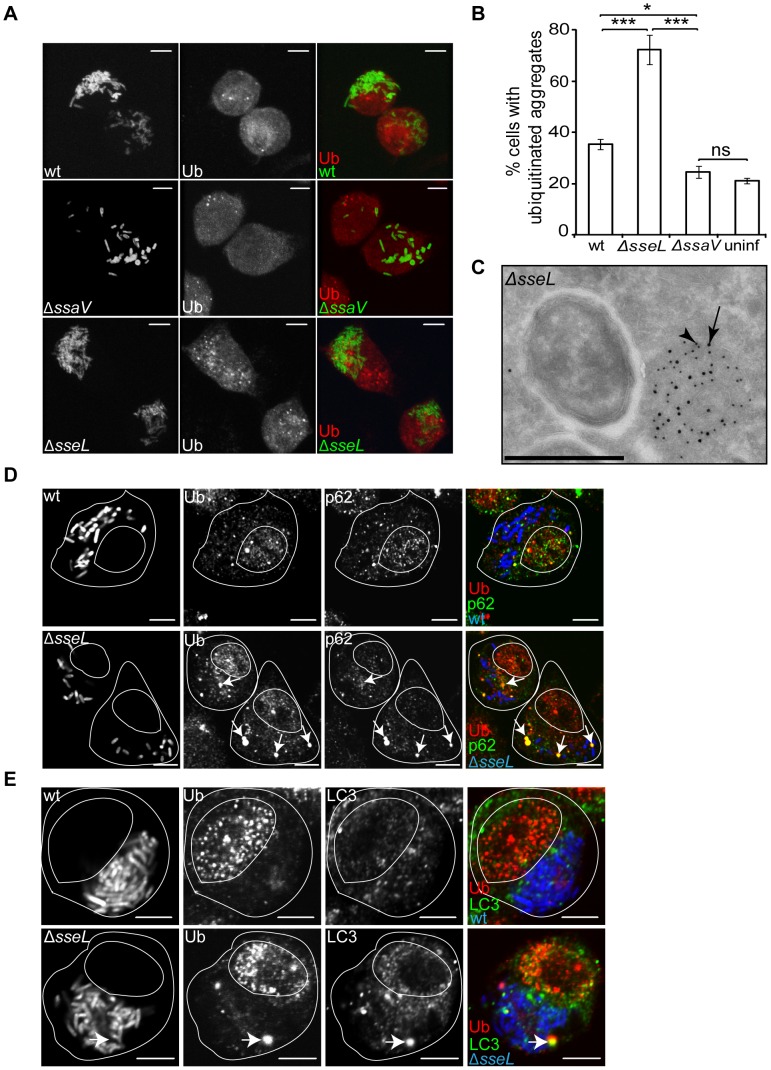
Figure 5. SseL reduces the accumulation of ubiquitinated aggregates and ALIS in macrophages.
(A) Single confocal sections of RAW264.7 macrophages infected with GFP-expressing strains of S. Typhimurium (green) for 10 h and immunolabelled for ubiquitin (Ub, red) (scale bars, 5 µm). (B) Quantification of infected macrophages containing ubiquitinated aggregates 10 h after bacterial uptake. Cells were processed as in (A) and analysed by fluorescence microscopy. A minimum of 50 cells were counted for each bacterial infection per experiment and values are the mean ± SEM of at least 3 independent experiments. * p<0.05; *** p<0.001. Uninfected cells (uninf) were from the same wells as infected and therefore were exposed to extracellular bacteria. (C) Immunoelectron microscopy of RAW264.7 macrophages infected with the ΔsseL mutant bacteria for 12 h. Arrows and arrowheads indicate ubiquitin (15 nm gold particles) and p62 (10 nm gold particles), respectively (scale bar, 0.5 µm). (D and E) Single confocal sections of RAW264.7 macrophages infected with GFP-expressing strains of S. Typhimurium (blue) and immunolabelled for ubiquitin (Ub, red) and (D) p62 (green) or (E) LC3 (green). The far right panels show merged images of p62 or LC3, ubiquitin and Salmonella. Arrows indicate Arrows indicate SCV-associated ubiquitin labelling with p62 or LC3 (scale bars, 5 µm). Cell outlines and nuclei are delineated by white lines.