Abstract
Ubiquitination is a reversible protein modification that influences various cellular processes in eukaryotic cells. Deubiquitinating enzymes remove ubiquitin, maintain ubiquitin homeostasis and regulate protein degradation via the ubiquitination pathway. Cryptococcus neoformans is an important basidiomycete pathogen that causes life-threatening meningoencephalitis primarily in the immunocompromised population. In order to understand the possible influence deubiquitinases have on growth and virulence of the model pathogenic yeast Cryptococcus neoformans, we generated deletion mutants of seven putative deubiquitinase genes. Compared to other deubiquitinating enzyme mutants, a ubp5Δ mutant exhibited severely attenuated virulence and many distinct phenotypes, including decreased capsule formation, hypomelanization, defective sporulation, and elevated sensitivity to several external stressors (such as high temperature, oxidative and nitrosative stresses, high salts, and antifungal agents). Ubp5 is likely the major deubiquitinating enzyme for stress responses in C. neoformans, which further delineates the evolutionary divergence of Cryptococcus from the model yeast S. cerevisiae, and provides an important paradigm for understanding the potential role of deubiquitination in virulence by other pathogenic fungi. Other putative deubiquitinase mutants (doa4Δ and ubp13Δ) share some phenotypes with the ubp5Δ mutant, illustrating functional overlap among deubiquitinating enzymes in C. neoformans. Therefore, deubiquitinating enzymes (especially Ubp5) are essential for the virulence composite of C. neoformans and provide an additional yeast survival and propagation advantage in the host.
Introduction
Cryptococcus neoformans is a very important basidiomycete fungal pathogen, which can cause life-threatening meningoencephalitis primarily among immunocompromised hosts. It has been estimated that there are over 1 million new cases of cryptococcal infection in HIV-infected patients each year, and approximately 625,000 deaths are associated with cryptococcal meningoencephalitis [1]. Furthermore, cryptococcosis has also become one of most common invasive fungal infections in patients with solid organ transplantations or other immunosuppressive conditions [2]. Finally, an outbreak of cryptococcal disease among immunocompetent humans and animals on Vancouver Island and the Pacific Northwest, USA since 1999 caused by Cryptococcus gattii (a species that is closely related to C. neoformans) emphasizes that this infection can break out of the classic opportunistic risk groups [3]. Undoubtedly, cryptococcosis has become a major pathogenic burden on human health.
Cryptococcus can overcome various hostile conditions in vivo (such as high temperature, oxidative and nitrosative stresses, nutrient starvation, hypoxia, and high CO2) to produce disease [4], [5]. Several pathogenic factors contribute to the rapid adaptation of Cryptococcus neoformans inside the mammalian host and have been genetically characterized, including its polysaccharide capsule [6], melanin [7], ability to grow at 37°C [8], urease [9], and phospholipase [10] . In addition, substantial progress has been made in understanding the signal transduction pathways regulating virulence and sexual reproduction over the last decade [11], [12], [13]. Due to its well-defined sexual cycle, robust animal models and molecular biology tools, C. neoformans has become an excellent model for the study of fungal pathogenesis [14], [15]. However, these advances have confirmed the fact that fungal virulence is a very complicated phenotype. Identification of the essential complex regulatory mechanisms in control of the cryptococcal virulence composite and its networks will be helpful in identifying targets to abrogate disease caused by C. neoformans.
Ubiquitination is an essential posttranslational modification of cellular proteins in eukaryotic cells, which marks certain proteins for degradation by the proteasome and thus is a major regulator of protein activity and/or localization. Besides ubiquitin and the proteasome, the ubiquitin pathway also includes multiple enzymes such as E1 (ubiquitin activating enzyme), E2 (ubiquitin conjugating enzyme), E3 (ubiquitin ligase), and the DUBs (deubiquitinating enzymes or deubiquitinase) [16], [17]. Ubiquitination comprises a series of reactions, in which ubiquitin is activated first by E1, transferred to E2, and finally recognizes and tags specific substrate proteins for degradation via the proteasome with the help of E3. DUBs not only process the ubiquitin precursors into mature monomers, but also help to re-utilize ubiquitin by cleaving it from the substrate before targeted protein degradation. The ubiquitin-proteasome system plays a central role in a variety of cellular processes such as cell cycle progression, signal transduction, transcriptional control, DNA repair, and stress responses [17], [18].
DUBs belong to a highly-conserved protease superfamily that can be divided into five subclasses based on their catalytic domain structure: Ubiquitin C-Terminal Hydrolases (UCHs), ubiquitin-specific proteases (USPs), ovarian tumor proteases (OTUs), JAB1/MPN/Mov34 metalloenzymes (JAMMs) and Machado-Joseph disease proteases [19], [20]. A large number of DUB genes have been discovered in eukaryotes (including approximately 19 in S. cerevisiae, 22 in Schizosaccharomyces pombe, and 95 in the human genome), thus demonstrating their essential functions [20], [21]. In addition to maintaining ubiquitin homeostasis, DUBs also play important roles in proofreading some specific ubiquitinated substrates and rescuing them from proteolysis or the non-degradative pathway [19].
In the last few years, significant progress has been made in understanding the structures and biological functions of DUBs in S. cerevisiae, S. pombe and humans [22], [23], [21]. In S. cerevisiae, DUBs participate in regulating energy metabolism, sexual reproduction, as well as various stress responses [24], [25], [26], [27], [28]. For example, Doa4 and Ubp10 regulate the utilization of carbon and nitrogen sources, respectively [24], [27]. Deletion of UBP14 leads to decreased meiotic efficiency, and a doa4Δ mutant shows a sporulation defect [25]. Most DUBs in S. cerevisiae are generally involved with the regulation of different stress responses [26], [29], [28]. It is noteworthy that the DUB-encoding gene UBP15 not only regulates the stress response due to temperature changes and chemical stresses, but also plays important roles in the growth rate and cell size of S. cerevisiae [29], [30], [31]. Recently, studies have suggested that DUBs take part in pathogenesis of C. neoformans via regulating various stress responses and melanization [32], [33]. However, the networks and mechanisms of deubiquitinating enzymes as major controllers of the fungal virulence composite remain to be further understood.
In this study, we use C. neoformans as a model to investigate the role of the deubiquitinating enzyme family on fungal pathogenesis. We found that deletion of the putative DUB-encoding gene UBP5 resulted in severely attenuated virulence and many distinct phenotypes, encompassing not only the defects in the classical pathogenicity factors such as capsule production and melanin synthesis, but also elevated sensitivity to several other important biological stresses and showed slower growth. In addition, we also confirmed that Ubp5 influences the sexual reproduction of this yeast. Our work illustrates the central importance of UBP5 in adapting to hostile growth environments including infection sites in the host, and highlights the divergence of this yeast from other eukaryotes in the evolution of the DUB enzymes.
Results
Identification of DUB Homologues in C. neoformans
In order to identify genes important for maintenance of infection and survival in the central nervous system (CNS), we performed comparative transcriptional profiling experiments using WT strain H99 and previously described mutant strains during growth in ex vivo cerebrospinal fluid (CSF) and serum, and during human or rabbit infection. As shown in Table S2, 17 genes putatively involved in the ubiquitin-proteasome system were identified as being differentially expressed in these conditions. Therefore, to appreciate the function of this in vivo regulated ubiquitination system, we focused on the importance of deubiquitinating enzymes (DUB) in C. neoformans disease.
We identified 19 putative DUB proteins in the C. neoformans strain H99 genome belonging to four of five DUB subfamilies (UCH, USP, OTU, and JAMM) by homology to known DUBs from S. cerevisiae and S. pombe. Of these, 12 have been previously described via signature-tag mutagenesis and their roles in Cryptococcus pathogenesis have been ascertained [32]. We deleted two of these putative virulence-related DUB genes, DOA4 and UBP14, in the WT strain H99 for use as controls in this study. Five of the remaining seven uncharacterized putative DUBs were deleted in the current study: Ubp5 (CNAG_05650), Ubp7 (CANG_03872), Ubp13 (CANG_01377), Otu1 (CANG_06428) and Rpn11 (CANG_07028). Of the two remaining genes (UCH2, CNAG_00989; UBP10, CNAG_01614), one (UCH2) may be an essential gene as we are unable to obtain a null mutant strain after numerous attempts. In contrast, putative null mutants of UBP10 exhibit unusual cell morphology and have not been further characterized.
Bioinformatic analysis of the putative cryptococcal DUB proteins in this study showed that five of them belong to the USP subfamily (the largest one in C. neoformans includes 15 DUBs), while the other two belong to either the OTU or JAMM subfamilies. Many of these DUBs have amino-terminal extensions while some have carboxyl-terminal extensions (Figure 1A). Even within the same subfamily, their catalytic domains exhibit significant differences in length, suggesting that there must be different insertions in the catalytic domains of many isoforms. Previous studies have confirmed that DUBs do display specificity for both substrates and particular ubiquitin chain types, yet they all share the basic role of cleaving off ubiquitin from other adduct or fusion proteins [34], [20], [21]. These extensions and insertions may contribute to the functional diversity of the DUB enzymes. Interestingly, we found that deletion of UBP5 generated many intriguing phenotypic changes in C. neoformans that were linked to virulence. Therefore, we focused in detail on UBP5 for its relationship to pathogenesis.
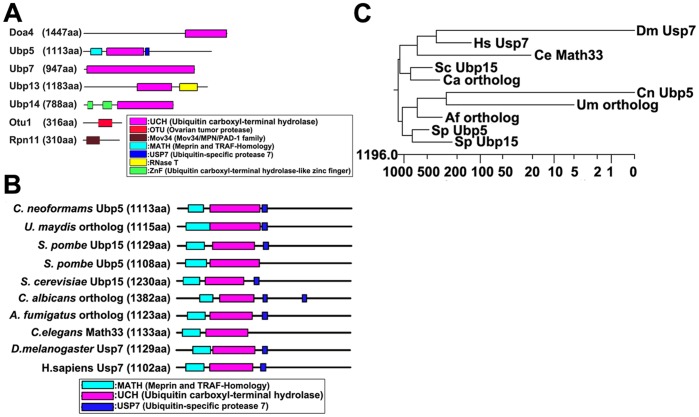
Figure 1. Structure analysis of deubiquitinating enzymes.
A. Domain structure of cryptococcal deubiquitinating enzymes. 7 DUB enzymes in this study belong to three subfamilies (USP, OTU, and JAMM) on the basis of their catalytic domains. USP and OTU domain DUBs are cysteine proteases, JAMM domain DUB is metalloprotease. We retrieved domain architectures for each DUB using the Pfam databases (http://pfam.sanger.ac.uk/). B. Comparison of Ubp5 orthologs between C. neoformans and other species. Each Ubp5 ortholog diagram shows functional protein domains, which were identified by the Pfam database. C. Phylogenetic tree analysis of Ubp5 orthologs is depicted by Clustal W alignment from DNASTAR software (versions 6.13). Protein sequences of Ubp5 orthologs were retrieved from the following database: C. neoformans Ubp5 (CNAG_05650) from the C. neoformans var. grubii H99 database of the Broad Institute, and its orthologs among other eukaryotic species from the protein databank of NCBI website. Cn, Cryptococcus neoformans; Um, Ustilago maydis; Sp, Schizosaccharomyces pombe; Sc, Saccharomyces cerevisiae; Ca, Candidia albicans; Af, Aspergillus fumigatus; Ce, Caenorhabditis elegans; Dm, Drosophila melanogaster; Hs, Homo sapiens.
We compared protein sequences of CNAG_05650 and Ubp5 orthologs from different eukaryotic species. Sequence analysis indicated that CNAG_05650 encodes 1113 amino acids in multiple domains, including an N-terminal MATH domain (the Meprin And TRAF-Homology domain, E-value: 3.4e-06), a UCH domain (Ubiquitin carboxyl-terminal hydrolase, E-value: 3.8e-59), and a C-terminal USP7 domain (Ubiquitin-specific protease 7, E-value:2.4e-12) (Figure 1A). Domain arrangement was similar to S. cerevisiae deubiquitinating enzyme Ubp15 with 38%, 54% and 44% identity in the three domains. An alignment of C. neoformans CNAG_05650 with orthologs from other eukaryotic species revealed conservation in protein domain structure (Figure 1B & 1C).
Characterization of DUB Roles in Stress Responses
To investigate the roles of deubiquitinating enzymes in growth and virulence of C. neoformans, seven DUB genes were deleted in the serotype A strain H99 (Table 1). Targeted disruption was confirmed by PCR and Southern hybridization. Subsequently, we reconstituted the ubp5Δ mutant to WT via ectopic integration of the WT gene, and all the phenotypes shown for the ubp5Δ mutant were complemented in the ubp5Δ+UBP5 reconstituted strain.
Table 1. Strains and plasmids used in this study.
| Strain and Plasmid | Genotype | Reference |
| C. neoformans | ||
| H99 | MATα | [98] |
| KN99a | MATa | [101] |
| DF001 | MATαubp5::NEO | This study |
| DF002 | MATa ubp5::NAT | This study |
| DF003 | MATαubp5::NEO+ UBP5::NAT | This study |
| DF004 | MATαdoa4::NAT | This study |
| DF005 | MATαubp7::NEO | This study |
| DF006 | MATαubp13::NEO | This study |
| DF007 | MATa ubp14::NAT | This study |
| DF008 | MATαotu1::NEO | This study |
| DF009 | MATαrpn11::NEO | This study |
| DF010 | MATa::NAT | This study |
| TOC4 | MATαrim101::nat RIM101::NEO | [88] |
| ure1Δ | MATαure1::ADE2 | [9] |
| cap59Δ | MATαcap59::HYG | [101] |
| Plasmid | ||
| pJAF1 | NEO resistance gene | [88] |
| pCH233 | NAT resistance gene | [88] |
| pUBP5-NAT | Modified pCH233 containing gene UBP5 | This study |
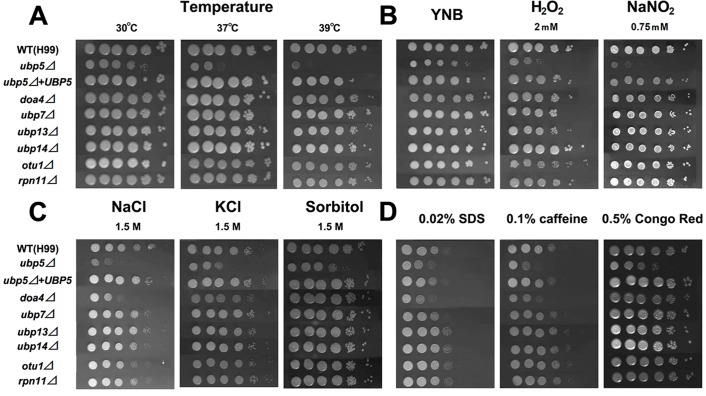
Since the ubiquitin-proteasome system plays important roles in regulating stress responses in eukaryotes [33], we tested the sensitivity of each mutant to a variety of in vitro stresses. Unlike the WT and other DUB mutant strains, only the ubp5Δ mutant exhibited temperature sensitivity (ts), with a partial growth defect at 37°C and complete growth inhibition at 39°C (Figure 2A). The ts phenotype was not rescued by the addition of 1 M sorbitol (data not shown) as was observed for a C. neoformans tps1Δ mutant that is also thermosensitive [35]. Moreover, deletion of UBP5 resulted in slower growth compared to WT even in rich medium at 30°C (Figure 2A and Figure S1). To examine whether this poor growth phenotype was due to carbon source preference, we also compared its growth using different carbon sources (YP agar medium with 3% galactose, 3% lactate, 3% glycerol, and 2% raffinose). No growth improvement was observed when the ubp5Δ strain grew in the media with alternate carbon sources (data not shown).
Figure 2. UBP5 is involved in various stress responses of C. neoformans.
A. Temperature sensitivity at 30°C, 37°C,39°C. B. Oxidative and nitrosative stress assay. C. High salts and osmotic stress assay. D. Cell-wall and cell-membrane stress assay. Strains were grown to saturation at 30°C in liquid YPD medium, 10-fold serially diluted (1–106 dilutions), and spotted 3 µl cells on YPD or YNB agar (containing different stress-inducing agents). Incubated them for five days and then photographed.
The ubp5Δ mutant also showed increased sensitivity to H2O2 (2 mM) and NaNO2 (0.75 mM), whereas the other DUB mutants exhibited no differences in sensitivity compared to the WT strain (Figure 2B). Likewise, the ubp5Δ mutant showed increased sensitivity to 1.5 M NaCl and 1.5 M KCl compared to WT, whereas the doa4Δ mutant exhibited increased sensitivity to elevated NaCl concentrations only (Figure 2C). Additionally, the ubp5Δ mutant showed increased sensitivity to 0.02% SDS, 0.1% caffeine or 0.5% congo red, whereas the doa4Δ mutant displayed increased sensitivity to 0.02% SDS and 0.1% caffeine (Figure 2D).
To further appreciate the impact of DUB enzymes on specific stress responses, all the mutants were exposed to various antifungal drugs: amphotericin B, caspofungin, fluconazole, or flucytosine. In vitro susceptibility testing was determined at 30°C according to the Clinical and Laboratory Standards Institute broth microdilution reference method (NCCLS, 2002). We observed that only the ubp5Δ mutant was hypersusceptible to all four antifungal agents compared with the WT strain, exhibiting an 8-fold reduction in MIC for flucytosine, 4-fold reduction in MICs for both fluconazole and caspofungin, and 2-fold reduction in MIC for amphotericin B (Table 2). The ubp5Δ+UBP5 reconstituted strain completely restored WT sensitivity to the four antifungal drugs. Among the other DUB mutants, both the doa4Δ and ubp13Δ mutants showed increased susceptibility to fluconazole and caspofungin, but not to amphotericin B or flucytosine.
Table 2. In vitro susceptibility of mutant strains to four antifungal agents.
| MIC80(µg ml−1) | ||||
| Antifungal compounds | Amphotericin B | Fluconazole | Flucytosine | Caspofungin |
| H99 | 0.5 | 8 | 16 | 32 |
| ubp5Δ | 0.25 | 2 | 2 | 8 |
| ubp5Δ+UBP5 | 0.5 | 8 | 16 | 32 |
| doa4Δ | 0.5 | 4 | 16 | 16 |
| ubp7Δ | 0.5 | 8 | 16 | 32 |
| ubp13Δ | 0.5 | 4 | 16 | 16 |
| ubp14Δ | 0.5 | 8 | 16 | 32 |
| otu1Δ | 0.5 | 8 | 16 | 32 |
| rpn11Δ | 0.5 | 8 | 16 | 32 |
| ATCC22019 | 0.5 | 2 | 0.125 | 0.5 |
The MICs of four antifungal agents against C. neoformans WT, DUB mutants and reconstituted strains were determined by the Clinical and Laboratory Standards Institute broth microdilution refercence method (NCCLS, 2002), and Candida parapsilosis strain (ATCC22019) was used as a quality control strain.
Following exposure to external stressors, misfolded or damaged proteins trigger activation of the ubiquitin-dependent proteolytic pathway. Maintenance of a sufficient ubiquitin pool is essential for this process, and ubiquitin homeostasis is at least partly maintained by the activity of DUB enzymes [19]. To understand the relationship between UBP5 deletion and ubiquitin stasis, we performed RT-PCR to examine the expression of UBI4 (encoding polyubiquitin) in both ubp5Δ mutant and WT strains. UBI4 expression was approximately 16-fold higher in ubp5Δ than in WT or reconstituted cells (Figure S3). Furthermore, deletion of UBP5 led to greatly elevated UBI4 expression at 37°C that peaked after 1 hr and then gradually declined. WT cells showed a gradual increase in UBI4 expression at 37°C but its peak value was still significantly lower than that in ubp5Δ cells, suggesting that UBP5 deletion may contribute to an increasing demand for polyubiquitin in C. neoformans cells to cope with high temperature and/or other stresses.
Taken together, deubiquitinating enzymes (especially Ubp5) appear to play important roles in regulating various stress responses. Deletion of Ubp5 made cryptococcal cells more susceptible to nearly all in vitro stresses that are closely related to pathogenesis compared to other DUB mutants. Therefore, we predicted that Ubp5 would play important roles in regulating pathogenesis of C. neoformans. Interestingly, phenotypes of several DUB mutants under stress conditions demonstrated both functional overlap and divergence within the deubiquitinating enzyme family for these pathogenic responses.
Roles of DUBs in Capsule, Melanin and Urease Production in C. neoformans
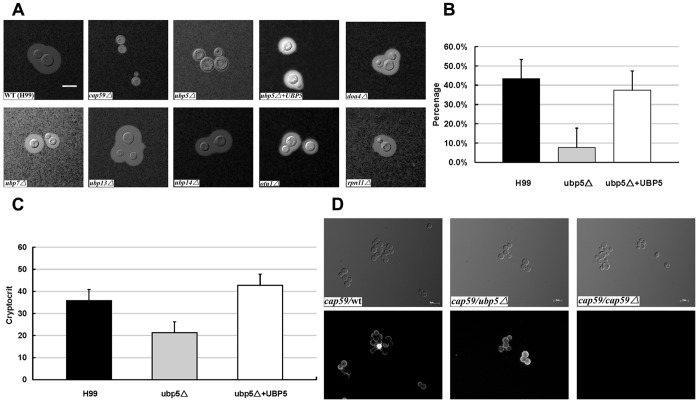
Several pathogenic factors are essential for C. neoformans to invade the host, such as capsule, melanin, and urease. When incubated under capsule-inducing conditions for 72 hrs, an obvious capsule defect was observed for the ubp5Δ mutant, whereas the reconstituted strain and other DUB mutants all showed WT capsule production (Figure 3A). In order to exclude the confounding effect of thermosensitivity, the ubp5Δ mutant was also examined in capsule-inducing medium at 30°C. Similarly, about 90% of ubp5Δ mutant cells displayed a significantly reduced capsule while 10% produced capsule similar to WT (data not shown). We measured the relative capsule volume (capsule diameter/total diameter) for the WT, ubp5Δ and ubp5Δ+UBP5 strains, and found that the volume of the ubp5Δ capsule was significantly smaller than WT (P<0.001) (Figure 3B). These data were confirmed using an independent measurement of capsule volume based on packed-cell volume (Figure 3C ).
Figure 3. Disruption of UBP5 significantly down-regulates the capsule production.
A. ubp5Δ mutants have a capsule defect. The WT strain H99 and all the DUB mutants were cultured on DME medium for capsule production at 37°C for 3 days. Capsule was assessed by staining with India ink and visualizing at 100 magnification (scale bar = 10 µm). B. Relative capsule volume detection. Total (cell and capsule) and cell-only diameter were measured via Photoshop Software for 50 cells for each strain. Then calculated the relative ratio of capsule with the formula ([Total Volume-Packed Volume]/Total Volume). After statistical analysis, the capsule production of ubp5Δ strain (7.7±0.48%) significantly decreased compared with either WT (43.4±6.41%) or reconstituted strain (37.4±8.01%) (P<0.001). C. Cryptocrit for Capsule Assay. After overnight growth, each culture was inoculated into tissue culture flasks with 20 mL DMEM medium in capsule-inducing condition for three days. Cells were killed with 10% formalin (V/V) for 5 minutes, and then counted. The samples were standardized to 3.2×108 cells/mL. The difference between ubp5Δ (21.28±3.00) and complemented (42.8±0.84) or WT (35.8±1.48) strains was P<0.001. D. Capsule transfer assay. Capsule material secreted from wild-type or ubp5Δ cells into conditioned medium (CM) can be attached to acapsular acceptor cells (cap59Δ). cap59Δ cell cannot shed capsule polysaccharide, thus its CM was used as a negative control. Cells were labeled with monoclonal antibody mAb18B7 and Alexa Fluror anti-mouse IgG (secondary antibody) was used for immunofluorescence.
Since C. neoformans capsule is excreted from the cell via exocytosis for attachment to the cell wall [36], a small proportion of ubp5Δ mutant cells with similar capsule like WT suggested that Ubp5 might not actually regulate capsule synthesis, but instead regulate attachment. Therefore, a capsule transfer assay was also performed [37]. Conditioned medium (CM) was prepared as a source of capsule polysaccharide by growing WT and ubp5Δ mutant cells in DMEM medium for one week; CM from acapsular cells (cap59Δ) was used as a negative control. Indirect immunofluorescence showed that acapsular cells could attach the capsular polysaccharide from the ubp5Δ CM medium (Figure 3D). Therefore, the ubp5Δ mutant synthesized capsule but was defective in extracellular assembly of the capsule on the cell surface.
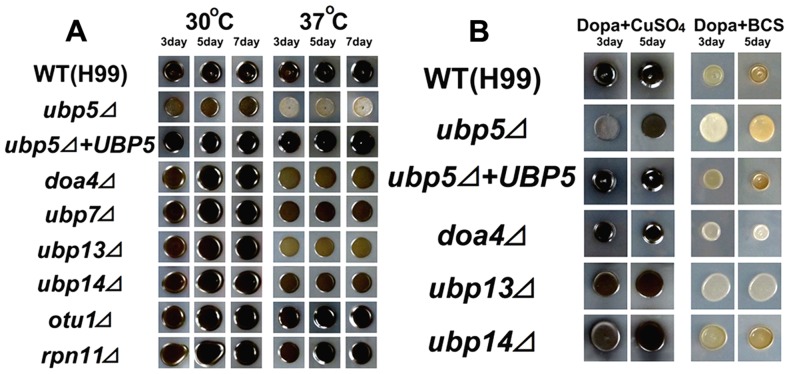
Melanin is another classic virulence factor in C. neoformans [38]. We assessed melanization for each of the DUB mutants on DOPA medium at both 30°C and 37°C. The ubp5Δ mutant strain exhibited hypomelanization on melanin-inducing medium at 30°C, and this defect was exacerbated at 37°C (Figure 4). However, hypomelanizaion in three other DUB mutants (doa4Δ, ubp13Δ and ubp14Δ) occurred at 37°C, suggesting that these deubiquitinating enzymes also influence melanization at mammalian host temperature. Copper availability is a major factor influencing melanization in C. neoformans. Loss of several genes related to copper ion metabolism (such as CCC2, ATX1 and CUF1) result in melanin defects in cryptococcal cells [39], [40], and many studies in Saccharomyces suggest that copper homeostasis is influenced by the ubiquitin system [41], [42]. Therefore, we hypothesized that DUBs might regulate melanization through their interaction with copper metabolism. We analyzed the melanization defect of the four DUB mutants by addition of exogenous CuSO4 and by chelation of copper ions from the media using bathocuproinedisulphonic acid (BCS). When copper ions were added into DOPA medium, melanin production was restored in all four DUB mutants (Figure 4B). However, after removing copper from DOPA medium, the doa4Δ and ubp13Δ mutant strains displayed completely non-melanized colonies whereas the ubp5Δ and ubp14Δ strains exhibited merely decreased melanization. Therefore, Ubp5 and Ubp14 may influence melanization by other mechanisms in addition to their influence on copper ion metabolism.
Figure 4. Melanin production assay of DUB mutants in C. neoformans.
A. Melanin production assay. Strains were grown on L-DOPA medium alone at 30°C and 37°C. B. Regulation of melanization by copper availability. Mutants strains with hypomelanization were incubated onto DOPA agar containing + 10 mM CuSO4, or containing + 1 mM BCS, a copper chelator at 37°C.
Urease is another important virulence factor of C. neoformans, which promotes increased CNS invasion and non-protective immune responses during lung infection [43], [44]. When incubated under Christianson’s urea agar, all the DUB mutants displayed WT urease activity as measured by the medium color change from yellow to bright pink (Figure S2). Therefore, these DUB enzymes do not influence urease activity.
Ubp5 is Required for Sexual Reproduction of C. neoformans
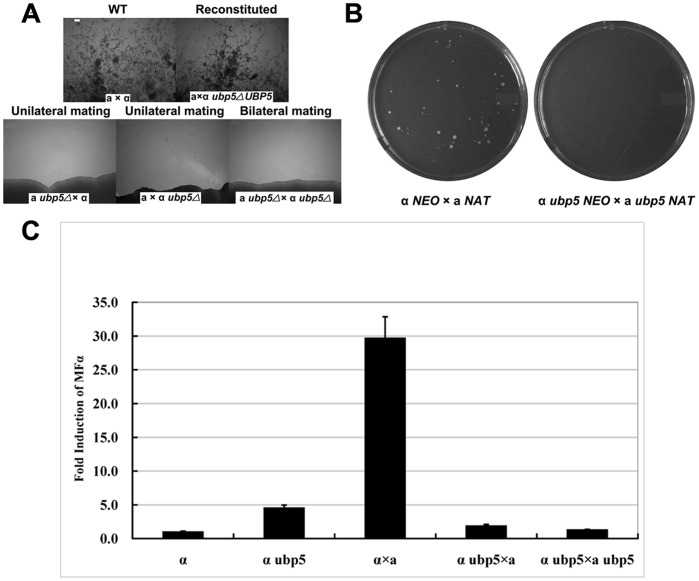
The UBP5 deletion was performed in both C. neoformans H99 (MATα) and KN99a (MATa) strain backgrounds. Sexual development and production of dikaryotic hyphae and basidiospores were examined in both unilateral crosses (ubp5Δ × WT) and bilateral crosses (MATa ubp5Δ × MATα ubp5Δ). Interestingly, no hyphae nor basidiospores were observed in unilateral crosses after two weeks co-incubation with mating partners (Figure 5A). In order to eliminate the effect of decreased growth rate on mating, we repeated the unilateral cross tests with a minor modification, mixing ubp5Δ and WT cells at 10∶1 ratio, and the result was still negative (data not shown) indicating that a single copy of UBP5 is insufficient to allow mating in C. neoformans.
Figure 5. C. neoformans Ubp5 is required for mating.
A. The ubp5Δ mutant is sterile in both unilateral and bilateral crosses. Reintroduction of the wild-type gene restores mating to a wild-type level. These fuzzy projections are mating-specific dikaryotic filaments. Mating assays were on V8 agar medium for 14 days at room temperature (scale bar = 10 µm). B. The ubp5Δ mutant is defective in cell fusion. Wild type cells (H99 and KN99a) with different resistance genes (NEO and NAT) were set as a control. The MATα and MATa cells were mixed for 24 (WT strains) or 72 hours (ubp5Δ mutant strains), serial diluted, and plated onto YPD argar containing both nourseothricin and neomycin. Following fusion, dikaryotic cells containing both wild-type MATα and MATa cells were able to grow. However, no viable dikaryotic cells were on the double selective plate of ubp5Δ mutant. C. MFα pheromone gene expression was assessed in a WT and ubp5Δ mutant strains alone, and in unilateral and bilateral crosses. Cells were incubated on V8 mating medium for 24 h at room temperature. RNA was extracted, and MFα gene expression was detected by real-time PCR with primers MFα-a/b (Table S1).
To identify where the defect in sexual reproduction occurred, we also performed a cell fusion assay. For this assay, strain DF001 (MATα ubp5Δ NEO) was mated with either strain DF002 (MATa ubp5Δ NAT) or DF010 (MATa NAT) on V8 agar for 24 h or 72 h and then plated on double selective medium (YPD containing neomycin and nourseothricin). We observed that ubp5Δ strains did not form colonies in bilateral crosses, suggesting Ubp5 is absolutely required for cell fusion (Figure 5B). We also tested fusion in unilateral crosses between ubp5Δ and WT; lack of viable colonies from this mating indicates that UBP5 in each mating partner is necessary for cell fusion (data not shown). This is consistent with cpk1Δ, ste7Δ, and ste11Δ mitogen-activated protein kinase (MAPK) pathway mutants, which also exhibit defects in fusion in unilateral crosses [45].
The MFα pheromone genes are important to initial cell fusion, and are induced by nutritional limitation and co-incubation with MATa cells [46]. MFα induction was assessed by real-time PCR in WTα and ubp5Δα mutant strains alone, and in mixed cell populations of unilateral and bilateral crosses. After 24 h co-culture with WT MATa cells, a dramatic induction of the MFα pheromone gene occurred in the WT MATα strain (Figure 5C) consistent with a previous report [46]. In contrast, when ubp5Δα mutant cells were co-incubated with WT or ubp5Δ mutant MATa cells, MFα expression showed no induction (Figure 5C). In addition, MFα transcription induced by limited nutrition alone was slightly higher in the ubp5Δ mutant than in WT. These results indicate that Ubp5 plays a complicated role in pheromone expression, which is not only required for induction of MFα transcription during mating but also negatively regulates its expression under nutrient-limited conditions.
Roles of DUBs on Cryptococcal Survival in Macrophages
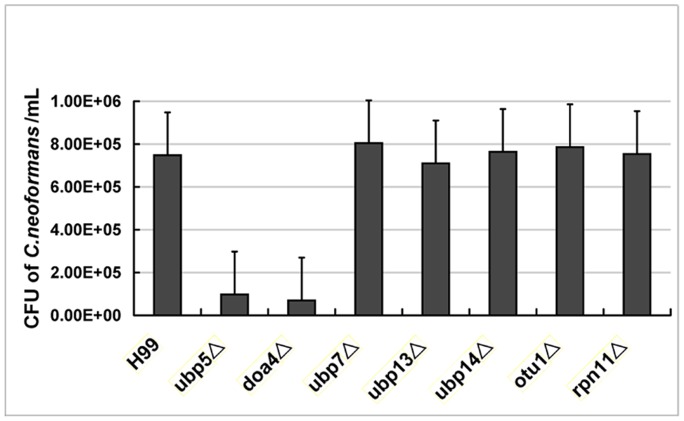
C. neoformans is a facultative pathogen, which can proliferate in both intracellular and extracellular environments [47]. Macrophages are primary professional phagocytes in the host, and also important for mediating the dissemination of cryptococcal cells from the lungs to the brain [48]. Therefore, we evaluated the DUB mutants for their ability to survive inside macrophages. The ubp5Δ and doa4Δ mutants exhibited significant reduction (87% and 92%, respectively) in their intracellular survival after co-inoculation with activated macrophages in DMEM for 24 hours, while other DUB mutants displayed WT survival (Figure 6). Interestingly, ectopic integration of UBP5 failed to restore intracellular survival in macrophages in spite of restoring all in vitro phenotypes tested.
Figure 6. Ubp5- and Doa4-mediated survival in macrophages.
Activated J774A.1 macrophages were co-incubated with the WT, all the DUB mutants and complemented strains of C. neoformans for 2 hours at 37°C in 5% CO2. Extracellular yeasts were then removed, and cocultures were incubated overnight under the same conditions. Following co-incubation, macrophages were lysed via 0.05% SDS and viable C. neoformans colonies were counted on YPD plates after 3–4 days’ growth at 30°C. Each strain was tested in triplicate (P ubp5 Δ = 0.003, P doa4 Δ<0.001).
Multiple virulence factors of C. neoformans contribute to its intracellular replication inside macrophages, including production of capsule, melanin, and a variety of secreted enzymes [49], [47], [50], [51]. Obviously, lower intracellular survival of the ubp5Δ mutant might result from a complex of several virulence-related phenotypic changes such as capsule and melanin defects, hypersensitivity to nitric oxide or hydrogen peroxide stress, slower growth rate, as well as decreased thermotolerance. However, the doa4Δ mutant exhibited significantly impaired intracellular survival despite displaying WT phenotypes for many of these virulence-related traits. Therefore, we hypothesize that Doa4 may regulate intracellular proliferation of C. neoformans by as yet additional undefined means.
Influence of Ubp5 on in vivo Virulence
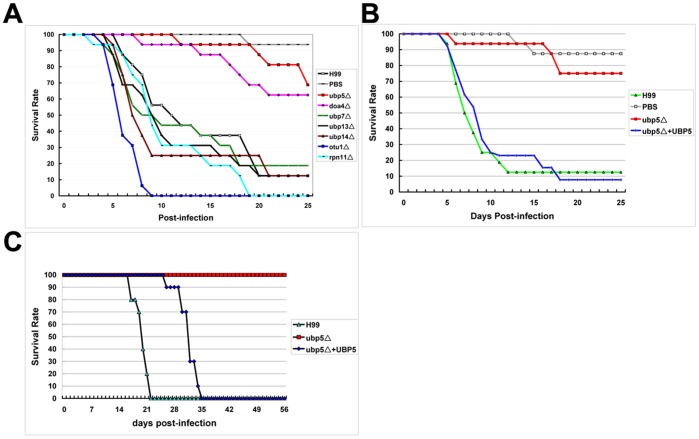
Galleria mellonella is a useful non-vertebrate model for high-throughput screening of in vivo virulence of C. neoformans [52], [53], [54]. In order to assess whether deubiquitinating enzyme deletions result in attenuated virulence, we employed a G. mellonella survival assay to assess our seven DUB enzyme mutant strains. Various degrees of virulence attenuation were observed in this invertebrate model (Figure 7A). The ubp5Δ and doa4Δ mutants showed a significant virulence attenuation: mean survival was 23.6±0.94 days for ubp5Δ (P<0.001) and 21.7±1.25 days for doa4Δ (P = 0.001) compared to 13.7±1.6 days for WT. In contrast, the otu1Δ mutant exhibited hypervirulence (6.4±0.36d, P<0.001) compared to WT strain (13.7±1.6d). None of the remaining mutants exhibited altered virulence compared to WT. These results are consistent with the macrophage killing assay, and both assays illustrated that different DUB enzymes impact discrete aspects of C. neoformans virulence despite their overarching functional similarities.
Figure 7. In vivo virulence assay of DUB mutants in G. mellonella and mouse infection models.
A. Screening assay of DUB mutant strains in the larvae model revealed various degrees of virulence. B. Reconstituted strain (ubp5Δ+UBP5) completely restored the virulence in G. mellonella killing assay. C. Deletion of Ubp5 exhibited significantly attenuated virulence in murine inhalation infection model.
The ability to survive at mammalian body temperature is required for pathogenicity of C. neoformans; thermosensitive strains are attenuated in the mammalian host. Since deletion of UBP5 impaired growth of C. neoformans at 37°C in vitro, we next assessed the virulence of the ubp5Δ mutant in a murine inhalation model of cryptococcosis. Female A/Jcr mice (10 per strain) were inoculated per nasally with 105 CFU of the wild-type, ubp5Δ mutant, or ubp5Δ+UBP5 reconstituted strains. Mice were monitored for survival and sacrificed at predetermined clinical endpoints. In contrast to the WT infected mice that exhibited complete mortality by 22 days post-inoculation, none of ubp5Δ-infected mice showed symptoms even after eight weeks post-infection, demonstrating a major virulence defect (P<0.001) (Figure 7C). Assessment of fungal burden in different organs showed that in comparison to WT or reconstituted-strain infected mice, viable cells were discovered from the lungs of mice infected by the ubp5Δ mutant but not the spleens or brains. Clearly, the ubp5Δ mutant is severely attenuated in the mammalian host for dissemination and disease even though the mutant exhibited prolonged survival in the lung. In contrast to the macrophage killing assay, reconstitution of UBP5 into the ubp5Δ mutant partially restored its virulence in mice while completely restoring the G. mellonella virulence and other in vitro phenotypes (Figure 7B and C).
UBP5 Deletion Effects on the C. neoformans Transcriptome
To further gain insight into the molecular basis for the pleiotropic phenotypes caused by UBP5 deletion, we carried out a microarray analysis of the differential genome-wide transcription profile of the ubp5△mutant versus its isogenic WT parental strain under both ambient and elevated temperature (37°C). These expression data have been submitted to the GEO database (http://www.ncbi.nlm.gov/gds) under accession number GSE35067. Among 10601 probes on the array, approximately 21% (1,463 genes) of the C. neoformans transcriptome showed significantly different expression patterns in the ubp5△mutant compared to the WT (P<0.05). Since the ubp5△mutant exhibited decreased melanin production at 37°C, the transcriptional change of the acid laccase-encoding gene CNM02420 (0.27-fold down-regulation, P = 5.82E-01) was selected as a reference cutoff. Approximately 653 genes were identified as biologically significant based on these criteria.
We specifically assessed how ubp5 mutation affects gene transcription pattern in C. neoformans at 30°C. Genes regulated by DUB enzyme Ubp5 deletion covered a wide variety of functional categories (Table 3). First, a group of genes involved in nutritional metabolism exhibited differential expression in the ubp5△ mutant compared to WT. Several enzymes in carbon utilization (e.g. sorbitol, alcohol and fatty acid) and ion stasis (e.g. sodium, calcium and iron) were up-regulated while a number of nitrogen-metabolite transporters (e.g. ammonium, spermine and urea) were repressed, suggesting that UBP5 deletion may alter metabolic flux in C. neoformans. Furthermore, several genes involved in stress defenses, such as CNAG_07836 (NAD binding dehydrogenase), CNL03740 (catalase), SOD2 (manganese superoxide dismutase) [55], AIF1 (apoptosis-inducing factor) [56] and QDR3 (Multidrug transporter of the major facilitator superfamily), were differentially expressed by UBP5 mutation. Ubp5 inactivation also resulted in many up-regulated genes associated with cell wall/membrane organization. This may partially explain the increased susceptibility to multiple cell-wall stressors and antifungal agents in the ubp5△ mutant (Figure 2 and Table 2). Finally, Ubp5 inactivation also led to differential expression of several genes involved in several basic cellular processes such as DNA replication, transcriptional regulation and cell cycle, and may explain the decreased growth rate at 30°C.
Table 3. Ubp5p-dependent gene expressions at 30°C.
| Category | Gene ID* | Fold change ubp5?/WT | P value | Functional annotation* |
| Carbohydrate/amino acid metabolism | CNAG_00269 | 1.80 | 8.30E-09 | Sorbitol dehydrogenase |
| CNC05840 | 1.50 | 4.60E-07 | Glycosyl hydrolase | |
| CNAG_06035 | 1.49 | 2.00E-08 | Alcohol dehydrogenase | |
| CNC01140 | 1.38 | 3.00E-10 | Lipase | |
| CNB00370 | 1.29 | 1.80E-07 | Fatty acid beta-oxidation-related protein | |
| CNE01100 | 0.71 | 1.50E-09 | long-chain-fatty-acid-CoA ligase | |
| CND00530 | 0.72 | 8.90E-07 | Urea transporter | |
| CNC06440 | 0.70 | 4.80E-08 | Inositol-3-phosphate synthase | |
| CNJ02210 | 0.63 | 7.10E-07 | Spermine transporter | |
| CNJ02220 | 0.59 | 1.70E-09 | Adenine nucleotide transporter, involved in beta-oxidationof medium-chain fatty acid | |
| CNJ01880 | 0.50 | 4.40E-09 | Ammonium transporter (AMT2) | |
| Cell wall/membrane organization | CNE03150 | 2.73 | 7.00E-12 | 1,3-beta-glucanase, involved in cell wall beta-glucanassembly (Exg1p) |
| CNE03240 | 2.54 | 2.00E-18 | Chitin synthase (Chs7p) | |
| CNAG_06031 | 1.82 | 2.00E-12 | Beta-glucan synthesis-associated protein | |
| CNAG_04270 | 1.54 | 1.80E-09 | SNAP receptor, mediate membrane fusion | |
| CNI02290 | 1.47 | 4.80E-08 | Polysaccharide synthase (Cps1p) | |
| CNF01800 | 1.45 | 4.70E-07 | Chitin deacetylase | |
| CNN00660 | 1.42 | 1.30E-08 | Glucan 1,3 beta-glucosidase protein putative | |
| Ion transport andmetabolism | CNL05450 | 2.30 | 8.00E-10 | Sodium, inorganic phosphate symporter |
| CNAG_04337 | 1.65 | 1.00E-14 | Cation diffusion facilitator | |
| CNK01330 | 1.53 | 4.00E-11 | Calcium ion transporter | |
| CND00150 | 1.52 | 5.40E-09 | Ferric chelate reductase (Fre7p) | |
| CNE04050 | 1.43 | 2.00E-12 | Iron ion transport-related protein | |
| DNA replication and repair | CNAG_04696 | 1.91 | 1.00E-13 | DNA clamp loader |
| CNI04060 | 1.38 | 5.00E-10 | Mitochondrial genome maintenance protein | |
| CNN01100 | 0.73 | 1.10E-06 | Telomere maintenance protein | |
| CNN01650 | 0.62 | 4.70E-08 | DNA dependent ATPase for DNA repair (Rad26p) | |
| Transcriptional regulation | CNK03060 | 1.43 | 8.40E-09 | C2H2 zinc finger protein Zas1 |
| CNAG_04403 | 1.43 | 9.80E-08 | CCCH zinc finger protein | |
| CNB03490 | 0.64 | 3.70E-08 | Transcriptional regulator | |
| Stress response | CNAG_07836 | 1.94 | 2.00E-12 | NAD binding dehydrogenase with oxidoreductase activity |
| CNAG_04521 | 1.47 | 3.70E-09 | Apoptosis-Inducing Factor (Aif1p) | |
| CNL03740 | 1.36 | 3.90E-07 | Catalase | |
| CNI01590 | 1.29 | 1.00E-08 | Manganese superoxide dismutase (SOD2p) | |
| CNL04910 | 0.66 | 1.10E-09 | Multidrug transporter of the major facilitator superfamily (Qdr3p) | |
| Signal pathway | CND06250 | 1.43 | 5.80E-06 | MAP kinase kinase kinase |
| CNAG_00802 | 1.39 | 3.90E-06 | Calcineurin-binding protein (Cbp1p) | |
| Cell cycle | CNG01990 | 1.62 | 8.00E-11 | G1/s-specific cyclin pcl1 (cyclin hcs26) |
| CNI01430 | 1.32 | 7.70E-06 | Cyclin-dependent protein kinase regulator | |
| Other | CNE02820 | 1.59 | 1.30E-09 | AFG1-family ATPase |
| CNAG_06009 | 1.53 | 1.10E-09 | Cyclohydrolase | |
| CNC02450 | 1.48 | 1.50E-06 | Water channel | |
| CNL04120 | 0.54 | 2.00E-10 | Adenosylmethionine-dependent methyltransferase |
Gene ID and functional annotation obtained from NCBI database (http://www.ncbi.nlm.nih.gov/) or Broad database (http://www.broadinstitute.org/annotation/genome/cryptococcus_neoformans/) with additional hand editing.
We also examined gene expression at 37°C. Several novel findings were obtained from this analysis (Table 4). First, more genes involved in stress responses, such as TPS2, CNB02790 (alpha, alpha-trehalase), GST3 and GST6 (glutathione S-transferase) were differentially expressed at 37°C compared to 30°C. Second, high temperature further inhibited some genes related to virulence factors such as CAP64 (capsular-related protein) and FET3 (acid laccase). In addition, a large number of genes involved in nucleotide metabolism, RNA processing and modification, ribosome synthesis and cell division exhibited significant reduction in ubp5△ at 37°C.
Table 4. Ubp5p-dependent gene expressions at 37°C.
| Category | Gene ID* | Fold change ubp5?/WT | P value | Functional annotation |
| Carbohydrate/amino acidmetabolism | CNI00320 | 1.99 | 1.10E-07 | Glyceraldehyde 3-phosphate dehydrogenase |
| CNE03150 | 1.53 | 7.60E-06 | Cellulase (167.m03461) | |
| CNA00470 | 1.52 | 1.10E-09 | Fructose-bisphosphatase (181.m07830) | |
| CNAG_02225 | 1.41 | 1.60E-08 | Cellulase | |
| CNF03760 | 1.40 | 2.30E-07 | Carboxypeptidase C | |
| CNN00510 | 1.37 | 2.50E-08 | L-fucose permease | |
| CNG01150 | 0.73 | 4.70E-06 | Spermidine synthase | |
| CNAG_01118 | 0.72 | 5.40E-08 | Amino acid transporter | |
| CNA08130 | 0.72 | 2.90E-08 | 2-hydroxyacid dehydrogenase | |
| CNL04780 | 0.64 | 1.90E-08 | Carbonic anhydrase protein | |
| CNJ00690 | 0.59 | 1.20E-09 | Uracil permease | |
| Nucleotide metabolism | CNC04630 | 0.71 | 1.80E-06 | Thymidylate kinase |
| CNE00200 | 0.69 | 9.80E-08 | Transferase purine nucleotide biosynthesis | |
| CNJ01230 | 0.64 | 1.30E-09 | Thymidylate synthase | |
| RNA processing and modification | CNF00090 | 0.70 | 7.00E-11 | rRNA modification-related protein |
| CNE01090 | 0.69 | 6.50E-09 | rRNA primary transcript binding protein | |
| CNH02690 | 0.68 | 7.30E-08 | ATP-dependent rRNA helicase (Spb4p) | |
| CNG03270 | 0.66 | 2.70E-07 | RNA-3-phosphate cyclase rRNA processing | |
| CNJ00110 | 0.65 | 4.70E-08 | rRNA (adenine-N6 ,N6-)-dimethyltransferase | |
| Ribosome synthesis | CNC05320 | 0.66 | 1.00E-10 | Mitochondrial 60 s ribosomal protein l38 (yml38) |
| CNB01260 | 0.64 | 4.30E-09 | Ribosomal large subunit biogenesis-related protein | |
| CNB05550 | 0.62 | 5.00E-11 | Cytosolic large ribosomal subunit protein | |
| Cell division and cell cycle | CNAG_06092 | 1.30 | 3.70E-07 | Cyclin |
| CNK02970 | 0.72 | 4.00E-06 | Mitotic chromosome condensation-related protein | |
| CNAG_02267 | 0.72 | 2.00E-10 | Spindle assembly checkpoint protein SLDB | |
| CNAG_00259 | 0.68 | 2.00E-10 | Bud site selection protein | |
| Cell wall/membrane organization | CNE01950 | 1.47 | 2.20E-08 | UDP-N-acetylglucosamine transporter |
| CNA06800 | 0.72 | 1.20E-07 | CAP64 gene product (Cas31p) | |
| CNC06200 | 0.70 | 4.90E-09 | Ergosterol biosynthesis-related protein | |
| Transcriptional regulation | CNAG_04457 | 2.02 | 1.30E-07 | Transcription factor activity |
| CNAG_02877 | 1.60 | 2.40E-07 | Sequence-specific DNA binding transcription factor activity | |
| CNC02990 | 0.63 | 8.10E-08 | DNA-directed RNA polymerase | |
| CNAG_06916 | 0.60 | 1.80E-09 | Zinc finger protein | |
| Stress response | CNAG_03765 | 1.37 | 6.00E-09 | Trehalose-phosphatase (Tps2p) |
| CNA07730 | 1.34 | 2.30E-09 | Multidrug resistance protein 1 | |
| CNAG_01893 | 1.33 | 2.60E-07 | Glutathione S-transferase 3 (Gst3p) | |
| CNB02790 | 1.30 | 5.60E-09 | Alpha,alpha-trehalase | |
| CNM02420 | 0.73 | 5.82E-01 | Acidic laccase (Fet3p/5p) | |
| CNF02230 | 0.57 | 2.00E-11 | Glutathione S-transferase 6 (Gst6p) |
The table does not list those differential genes overlapped with Table 3.
Discussion
Modification of proteins by ubiquitination is a critical regulatory mechanism for various cellular processes. Proteomic studies indicate that about 1/5?1/6 of all proteins (1,075) are modified by ubiquitination in Saccharomyces [57]. Deubiquinating enzymes (DUBs), which mediate the removal and processing of ubiquitin from ubiquitin precursors, ubiquitin-protein conjugates, and unanchored ubiquitin chains, play essential roles in protein degradation by either the proteasome or other ubiquitin-dependent processes for their help in recycling ubiquitin. A previous cryptococcal study described the phenotype of 12 DUB gene deletion mutants regarding the classic pathogenicity factors (capsule production, melanin and high temperature growth) and in vivo virulence in the CMO18 strain background [32]. On the basis of that study and our own in vivo transcriptional analyses that suggested importance of this pathway for the virulence composite, we performed a comprehensive functional analysis of the DUB family in the model pathogenic yeast C. neoformans. C. neoformans possesses 19 putative DUB genes among which most appear to belong to the USP subfamily. In this report, we disrupted five of the remaining deubiquitinase genes in the H99 strain background, along with the creation of two confirmed virulence-related mutant strains (doa4Δ and ubp14Δ) in same background for use as controls [32]. None of these genes were essential and most DUB mutants did not exhibit significant growth defects under standard growth conditions. However, deletion of UBP5 led to severely altered phenotypes, which illustrates that this deubiquitinating enzyme plays a vital but specific role in regulating cell propagation, sexual reproduction, stress response and pathogenesis of C. neoformans.
The first distinct phenotype in the ubp5△mutant is its poor growth performance even in rich medium (Figure 2A) and alternative carbon sources do not improve its growth (data not shown). This observation may be partially contributing to the disordered nutritional utilization caused by Ubp5 deletion (Table 3). Some deubiquitinating enzymes in Aspergillus nidulans and S. cerevisiae also play roles in regulating their utilization of carbon resources [58], [27]. On the other hand, the growth suppression of ubp5△ mutant is associated with its decreased proliferation rate (Figure S1), which is similar to the null mutant of UBP15 (the orthologue of CnUBP5) in S. cerevisiae [59]. Through physical interaction with APC/C (anaphase-promoting complex/cyclosome, an ubiquitin-ligation enzyme) activator Cdh1p, ScUbp15p can reverse the degradation of cell cycle proteins via ubiquitination and drives the transitions through the different stages of the cell cycle [60], which then governs the rate of cell proliferation in S. cerevisiae [61]. In C. neoformans, Ubp5 inactivation contributed to differential transcription of several genes involved in cell cycle regulation. Therefore we hypothesize that Ubp5 may regulate cell division in a similar fashion in C. neoformans. The ubp5△mutant may be dependent on such regulatory mechanisms to reduce non-essential energy consumption and to meet enhanced energy demand due to high temperature, since Ubp5 deletion may disturb normal energy mobilization and utilization in C. neoformans (Table 3).
In addition to asexual propagation, we found that Ubp5 also plays an essential role in sexual reproduction and possibly production of one of the primary infectious propagules (basidiospores) in human cryptococcosis [62]. Sexual development of cryptococcal cells involves a series of fundamental events such as plasmogamy, karyogamy, meiosis and sporulation [63]. Our tests showed that the mating of ubp5Δ mutant with WT fails to produce any filaments, and appears to be blocked at the point of cell fusion. This is partially attributed to blocked MFα pheromone gene induction during mating (Figure 5). A recent study illustrates that the ubiquitin ligation enzyme Fbp1 also regulates sporulation in fbp1Δ bilateral crosses of C. neoformans [64]. However, it remains to be elucidated whether it works in concert with other genes such as the DUBs. Several ubiquitin-related proteins, such as Ubi4 (polyubiquitin), Doa4, and Ubp14 (deubiquitinating enzymes) were confirmed to function in the coordination of sexual reproduction in S. cerevisiae, where deletions of these genes lead to a sporulation defect [65], [25]. The pathway may be dependent on the degradation of MAPK pathway components or the transcriptional repressor of mating-type specific genes mediated by ubiquitination [66], [67], [68]. The ubiquitin pathway may also regulate sporulation in a similar way in C. neoformans. However, no sporulation defects have been shown in the ScUbp15 mutant as have been observed for the C. neoformans orthologue CnUbp5. This illustrates the functional divergence and redistribution among the deubiquitinating enzymes from different fungal species.
C. neoformans is a globally distributed and free-living pathogenic fungus with tenacious vitality, which is illustrated by its adaptation to dramatically different environments during infection within a mammalian host. Among many in vivo stressors for this yeast, increased environmental temperature is a major challenge that C. neoformans encounters upon initial entry into the host. We demonstrated that disruption of the C. neoformans UBP5 gene confered a conditional temperature-sensitive growth defect. Reconstitution of the wild-type UBP5 allele fully restored the ability of the ubp5Δ mutant to grow at 37°C. Sorbitol and trehalose are important sugars for thermotolerance in yeasts with their known ability to stabilize proteins, cell walls, and membranes [69], [35]. However, sorbitol did not suppress the ts phenotype of ubp5Δ, and the ubp5Δ mutant produced WT trehalose levels (data not shown). Therefore, the influence of Ubp5 on heat tolerance is likely independent of the trehalose pathway. Dozens of genes and several signaling pathways (such as MAPK, Ca2+/calcineurin, and Ras1 pathways) have also been identified as closely related to thermotolerance in C. neoformans [8], [70], [13]. For example, several mutants in the Ras1 pathway show a similar conditional ts phenotype, where the mutants are viable but fail to grow at 37°C or 39°C [71], [70], [72]. It remains to be proven whether Ubp5 is connected to the Ras1 pathway in regulation of thermotolerance in C. neoformans. Interestingly, no other deubiquitinating enzymes in C. neoformans appear to take part in regulation of thermotolerance other than Ubp5 [32]. In contrast, studies in S. cerevisiae reveal that several DUBs such as Doa4, Ubp8, Ubp10 and Ubp15 play roles in thermotolerance [22], and again illustrate both the functional similarities and divergences of DUBs between these two fungal species.
In addition to regulating thermotolerance, Ubp5 is also essential for the resistance of C. neoformans to other important biological stresses such as oxidative and nitrosative stress, salt stress, cell wall and cell membrane damaging agents, and antifungal agents (Figure 2 and Table 2). The ubiquitin-proteasome system has been identified as one of four core protein families in response to H2O2 stress across multiple eukaryotic species, including humans, plants, and both fission and budding yeasts [73]. A recent study demonstrated that the ubiquitin system is involved in both oxidative stress responses and in responses to antifungal agents such as fluconazole and amphotericin B in C. neoformans [33]. Consistent with these general phenotypic observations, our findings identified Ubp5 as specifically important for enhanced susceptibility to all four classes of antifungal agents, and suggest that a single deubquitinating enzyme may be implicated in the regulation of ergosterol synthesis, DNA/RNA biosynthesis and β-glucan formation in C. neoformans, which is also supported by our transcription profiling data (Tables 2 & 3). Along with Ubp5, we observed that the doa4Δ mutant also showed decreased resistance to high sodium ions, 0.02% SDS and 0.1% caffeine, and both doa4Δ and ubp13Δ mutants are more sensitive to fluconazole and caspofungin, demonstrating some functional overlap of the deubiquitinating enzymes in C. neoformans.
Presumably, various extracellular or intracellular stresses such as high temperature, oxidative and nitrosative damage, high salt, and antifungal drugs, lead to accumulation of misfolded or damaged proteins inside the yeast cell, and cause cell damage that is prevented by their removal through the ubiquitin-dependent proteolytic pathway. The importance of ubiquitin, as an essential element during stress responses, is further highlighted by the requirement of a functional polyubiquitin gene to withstand heat stress, elevated H2O2, and other stress agents in yeast [65], [74], [75]. Conversely, deubiquitination is not an essential pathway for general survival. The primary function of DUB enzymes is to maintain ubiquitin homeostasis via processing or recycling ubiquitin from its precursors or adducts [19]. Ubiquitin is likely encoded by two genes (UBI4 and UBI1) in C. neoformans, which produce a linear peptide consisting of polyubiquitin (five copies) or monoubiquitin fused to a ribosomal protein [76]. Therefore, we hypothesize that Ubp5 in C. neoformans may be the major deubiquitinating enzyme to deal with stress. We suspect that it works synergistically with Ubi4 to maintain ubiquitin homeostasis especially under stressful conditions (Figure S3) and these specific regulatory connections will need to be further understood.
Several studies confirm that signaling pathways, such as PKC/MAPK, HOG, and Ca2+/ calcineurin pathways, jointly coordinate the stress responses in C. neoformans [4], [77], [13] and studies in other eukaryotic species reveal that the ubiquitin-proteasome system plays an important role in the regulation of these signaling pathways [78], [79], [80], [81]. For example, the ubiquitin system can down-regulate the HOG pathway via selective degradation of the upstream regulator Ssk1p in S. cerevisiae [78]. A recent report illustrates that the ubiquitin system may also play roles in osmotic stress, oxidative damage, and antifungal stress pathways by regulating the HOG pathway in C. neoformans [33]. Our microarray data also demonstrate that Ubp5 inactivation highly induces the transcription of several genes encoding MAPK and Ca2+/calcineurin signal pathway components such as MAPKKK and CPB1. Therefore, we hypothesize that there is complicated crosstalk between deubiquitinating enzymes and other signaling pathways in C. neoformans that require further study to understand their total impact on the pathobiology of C. neoformans.
In this study, we performed biological screening with both a macrophage-killing assay and a G. mellonella infection model. These models assess different aspects of cryptococcal pathogenesis and allow us to examine the virulence composite differences of each DUB mutant in a biological context (Figure 6 and Figure 7A?B). We found that deletions in either the UBP5 or DOA4 led to dramatic attenuation in virulence in both models and there were no significant virulence differences between ubp5Δ and doa4Δ mutants in either model, although disruption of UBP5 did yield more altered in vitro phenotypes under several conditions. Reduced survival in the macrophage assay for both ubp5Δ and doa4Δ was observed, however, the reconstituted ubp5Δ+UBP5 strain was unable to restore survival for the ubp5Δ mutant in this assay. This result may be due to the ectopic integration of UBP5 in this strain suggesting that precise transcriptional control of UBP5 is required; alternatively, a secondary mutation may exist in this strain.
Our results showing attenuated virulence of the doa4Δ mutant in C. neoformans was consistent with a prior report on this mutant in the mouse inhalation model [32]. It should be noted that the doa4Δ mutant was not complemented in this prior work, nor in this current report. It should also be pointed out that the ubp14Δ mutant (in strain H99) displays wild-type virulence in our assay (P = 0.995) while the same mutant in the gene-deletion library (in strain CMO18) showed decreased lung infectivity in the mouse model [32]. These contradictory results may be attributed to differences between vertebrate and non-vertebrate hosts, and/or the known variation of virulence between the different parental strains.
We also used the mouse inhalation model to test the virulence of the ubp5Δ mutant. All the mice infected by the ubp5Δ mutant survived for 8 weeks with low burden of yeasts in lung tissue and no viable yeast cells in CNS. With the long-term yeast survival in lung tissue of mice without dissemination and attenuation in the room temperature heterologous host model (G. mellonella), clearly, thermotolerance is not the only virulence factor influenced by Ubp5.
C. neoformans virulence has been linked to several other well-defined factors, including the production of capsule, melanin, and various lytic enzymes [82], [83], [84]. Of these classical virulence associated traits, capsule attachment was altered in the ubp5Δ mutant (Figure 3). Interestingly, a small proportion of mutant cells (10%) still exhibited normal capsule production although most showed an apparent capsular defect. Capsule production is a complex and dynamic process including intracellular polysaccharide synthesis, secretion of capsular polysaccharide via vesicles, and extracellular assembly [85]. Our capsule assay indicates that UBP5 deletion does not affect capsule synthesis or secretion (Figure 3D), therefore Ubp5 probably participates in the polysaccharide assembly on the C. neoformans cell wall. Previous studies have identified that cell wall components such as alpha (1–3) glucan and chitin-derived molecules are required for GXM (major capsule component) anchoring to the cell surface [37], [86], [87]. In fact, enhanced susceptibility to cell wall-damaging agents such as caspofungin in the ubp5Δ mutant provides indirect evidence for its impact on cell wall formation. It has been reported that deletion of the transcriptional regulator RIM101 produces a similar capsule defect by altering the expression of genes responsible for capsule attachment to the cell wall [88]. However, we examined the transcriptional expression of AGS1 (encoding alpha-1,3-glucan synthase, key enzyme cell wall synthesis) and no significant differences were observed between ubp5Δ mutant and WT strain. It is possible that Ubp5 may be participating in regulation or modification of chitin-related structures or cell-wall components other than alpha-1,3-glucan for capsule assembly, which is also supported by our transcriptome profiling analysis in the ubp5△ mutant. On the other hand, ubiquitination and deubiquitnation are essential post-transcriptional modifications for many protein targets. Ubp5 may regulate the synthesis or degradation of the proteins (including Ags1) essential for capsule attachment at the protein translation level.
Our melanization assay did demonstrate some functional crosstalk among deubiquitinating enzymes in C. neoformans. In our study, several deubiquitinating enzymes (such as Ubp5, Doa4, Ubp13 and Ubp14) influenced pigment production in C. neoformans by their common coordination of intracellular copper homeostasis (Figure 4). Disruption of these genes resulted in hypomelanized colonies of C. neoformans at 37°C, a phenotype that can be suppressed by additional exogenous copper ions. Similar results were reported with two other genes, CCC2 (copper transporter) and ATX1 (copper chaperone), both of which could mediate intracellular transport of copper ion to regulate laccase transcription [39] . The melanin defect was not as profound in the other DUB mutants at 37°C, and high temperature may disturb regulation of copper metabolism. However, melanization is impacted with the ubp5Δ mutant at 30°C and this may be related to its decreased growth rate. Our results with H99 stand in contrast to a previous study [32], where deletion of DOA4 or UBP14 led to a melanin defect in the CMO18 strain at 30°C. Melanization generally provides C. neoformans an added survival advantage against both environmental predators and the mammalian host immune system [89], [90], [7]. However, disruption of either UBP13 or UBP14 does not attenuate the virulence of C. neoformans in both macrophage and wax moth killing assays as the ubp5Δ and doa4Δ mutants do despite their effects on melanization. This finding suggests that hypomelanization may not be a critical factor for attenuated virulence in the ubp5Δ and doa4Δ mutants and specifically, Doa4 may depend on other unrecognized mechanisms to regulate pathogenesis in C. neoformans.
In summary, we have clearly demonstrated that deubiquitinating enzymes, especially Ubp5, are necessary for the efficient phenotypic expression of multiple virulence factors in the model fungal pathogen C. neoformans. These enzymes play pivotal roles in regulating propagation, sporulation, and stress responses. Their distinct roles further illustrate the evolutionary divergence of certain pathogenic fungi from the model yeasts such as S. cerevisiae, and also provide an important paradigm for further exploring the functions and mechanisms of deubiquitinating enzymes on the virulence of other fungal pathogens, such as Candida albicans, A. fumigatus, Ustilago maydis, and Magnaporthe grisea. Furthermore, the pleiotropic phenotypes of Ubp5 highlight its potential as an antifungal drug target. Indeed, inhibitors of some deubiquitinating enzymes including Usp7 (Ubp5’s homolog in human) are current viable targets for anticancer therapy [91]. Therefore, we propose that specific deubiquitinating enzyme inhibitors could provide similar benefits in the prophylaxis and treatment of fungal diseases.
Materials and Methods
Strains, Plasmids, and Media
The C. neoformans strains used in this study are listed in Table 1. Strains were cultured at 30°C on YPD agar medium, and selective media contained nourseothricin (100 mg L−1) and/or neomycin (G418 200 mg L−1). Capsule-inducing medium, urease test medium and DOPA were prepared as previously reported [92], [93]. Stress media were created by adding different stress-inducing agents into YNB agar medium (0.67% yeast nitrogen base without amino acids, 2% glucose) or YPD agar medium before autoclaving. V8 medium (pH = 5.0) was used for mating and cell infusion assays. Medium for J774 murine macrophages was prepared as previously reported [94].
Transcriptional Profiling
Transcriptional profiles were generated for WT and previously described mutant strains [95] from either ex vivo CSF and serum cultures or in vivo rabbit and human samples. In the comparison of CSF and serum exposure, WT strain H99 was incubated in two 50 mL YPD broth cultures for 24 hours at 30°C with shaking, washed with sterile distilled water, and resuspended in 50 mL sterile distilled water. The cultures were then split in half and pelleted by centrifugation, with half of the cultures resuspended in filter-sterilized human CSF or serum from anonymous human donors. These cultures were then incubated at 37°C with shaking, and harvested at either 1 or 24 hours post-resuspension in each biological fluid for RNA extraction. In the comparison of ex vivo CSF and rabbit infection, WT strain H99 was grown as described above, resuspended in ex vivo CSF for 24 hours, and harvested for RNA extraction. In the comparison of rabbit and human samples, New Zealand White rabbits were inoculated with WT strain H99 and harvested by intracisternal spinal tap on days 1 and 7 as previously described [95]. For the human sample, RNA was extracted from CSF obtained from a de-identified patient with cryptococcal meningitis as previously described [96]. RNA for all samples was obtained from lyophilized cells using the TriZol extraction protocol (Invitrogen, Carlsbad, CA) followed by purification using the Qiagen RNeasy kit (Qiagen, Valencia, CA) as previously described [96].
In the comparison between ubp5△ mutant and wild-type strain, both strains were inoculated in 50 mL YPD medium and grown to mid-log phase (OD600?1.5) at 30°C. The cultures were then split into two parts and pelleted by centrifugation. One part of the cultures resuspended in 25 mL fresh YPD medium and incubated for 1 h at 37°C. Following RNA extraction, microarray hybridizations and data acquisition were performed at the Duke Microarray Facility (http://microarray.genome.duke.edu/) according to their established protocols for custom spotted arrays. All transcription profiling data generated for this study have been deposited in the Gene Expression Omnibus, a MIAME-compliant database at the National Center for Biotechnology Information, under Accession # GSE35067.
Construction of DUB Mutant Strains and Reconstitution
Each DUB gene was disrupted in H99 and/or KN99a strains background by overlap PCR [97]. The purified PCR fragments were reduced to a 2 µl volume and precipitated onto 10 µl gold microparticles (0.6 µm; Bio-Rad) for biolistic transformation [98]. Stable transformants were selected by plating cells onto YPD medium containing either neomycin or nourseothricin. Mutants were screened by PCR using appropriate primers (Table S1). Positive transformants identified by PCR screening were further confirmed by Southern blot analysis.
To complement the ubp5Δ mutant, a genomic DNA fragment containing ORF, promoter and terminator region was amplified using primers Re0104-F and Re0104-R. This PCR fragment and the digested plasmid pCH233 by Xba I were fused with In-Fusion® EcoDry™ Cloning System (Clontech). The reconstructed vector pUBP5-NAT was reintroduced into ubp5Δ mutants via biolistic transformation. Positive colonies were selected on YPD agar plus nourseothricin. Reconstitution was confirmed by PCR and Southern hybridization.
Southern Blot Analysis
The mutant and reconstituted strains were reconfirmed using Southern hybridization analysis for gene replacement. Genomic DNA (20 µg) was digested with proper restriction endonucleases and separated on a 0.8% agarose gel. The DNA was transferred to a positively charged nylon membrane (Roche Applied Science, Indianapolis, IN). Membranes were hybridized with NEO, NAT or target-gene digoxigenin-labeled DNA probes overnight, as specified by the manufacturer (Roche Applied Science, Indianapolis, IN) washed and detected on the membrane [94]. DNA bands of interest were visualized on film following 3 to 10 min exposures.
RNA Extraction, cDNA Synthesis and Quantitative Real-time PCR
Following incubation to mid-log phase, 50 ml of C. neoformans cells were harvested by centrifugation at 1,800 ×g for 5 min, washed once with 1× PBS, and lyophilized overnight. The lyophilized pellet was then vortexed with 3 ml glass beads and resuspended in 4 ml TRIzol Reagent (Sigma). After sitting at room temperature for 5 min, 800 ml chloroform was added and the mixture was shaken for 30 s. The cell lysate was then centrifuged at 8,000 ×g for 10 min, and the supernatant was transferred to a new tube. Same volume of isopropanol was added, incubated for 10 min at room temperature, and centrifuged at 8,000 ×g for 10 min. After washing the pellet with 75% ethanol, it was diluted into 300 µl RNase-free water and incubated with DNase I at 37°C for 1 h. The RNA was purified with RNeasy Mini Kit (QIAGEN) again, and then stored at −80°C. First-strand cDNA was synthesized using random hexamer primers via the SuperScript III First-Strand Synthesis Kit for RT-PCR (Invitrogen) according to its protocol.
The resulting cDNA was used as template for quantitative real-time PCR using iQ SYBR Green Supermix (Bio-Rad) according to the manufacturer’s recommendations. Primers used for each gene are listed in Table S1 in the supplemental material. The multicolor real-time detection system was used with the following PCR conditions: an initial denaturing cycle of 95°C for 3 min and 40 cycles of denaturation at 95°C for 10 s and annealing/extension at 55°C for 20 s. Each reaction was performed in triplicate. The constitutively expressed GPD1 gene was used to normalize gene amplification for each strain and condition [99], and expression (n-fold) was calculated relative to the WT strain H99 using the Bio-Rad iCycler software system as previously described [93].
In vitro Stress Assays
Each strain was incubated to saturation at 30°C in YPD medium, washed, serially diluted (1 to 106dilutions) in 1× PBS, and spotted (3 µl) onto YNB or YPD agar medium containing different stress-inducing agents. For the oxidative and NO stress test, 2 mM H2O2 and 0.75 mM NaNO2 (pH = 4.0) were added into YNB agar medium. For the osmotic stress and high salt sensitivity test, we added 1.5 M sorbitol, 1.5 M NaCl, 1.5 M KCl to YPD agar medium. To examine cell wall integrity, the cells were spotted onto YPD agar media containing 0.02% SDS, 0.1% caffeine and 0.5% congo red. Then spotted cells were incubated at 30°C for 5 days and photographed.
All the mutant strains, along with WT, were analyzed for their sensitivities to the stress of antifungal compounds, such as amphotericin B, fluconazole, flucytosine and caspofungin. Tests were performed in accordance with NCCLS protocol M-27A [35], and Candida parapsilosis ATCC22019 was used as a quality control strain. The MIC80 was determined to compare the antifungal activity among different strains.
Assays for Melanization, Urease, Capsule Production
For the melanization test, cells were spotted onto L-DOPA agar and incubated for 3–7 days at 30°C and 37°C. Melanin production was monitored and photographed. For urease assay, strains were streaked onto Christiansen’s urea agar at 30°C for 24 hours and checked for the color change with WT and ure1Δ strains as controls [92].
For capsule production, different C. neoformans strains were incubated into DME medium or low iron medium at 37°C in 5% CO2 for 3 days in tissue culture flasks. Following capsule induction, the cells were washed once with 1× PBS, and resuspended in 1 ml sterile PBS for visualization of capsule. Each strain was mixed with India ink preparations on glass slides for visualization. Images were collected at a 100× magnification. Total (cell and capsule) and cell-only diameters were compared for 50 cells for each strain. Cryptocrit was also used to measure differences in capsule [93]. In order to examine the capsule secretion, a capsule transfer assay was performed per previous studies [37]. Briefly, conditioned medium (CM) was prepared as a source of capsule polysaccharide by growing WT and ubp5Δ mutant cells in DMEM medium for one week while CM from acapsular cells (cap59Δ) as a negative control, after which 2.5×106 of acapsular acceptor cells (cap59Δ) were incubated in the CM media. For indirect immunofluorescence, cells were co-cultured for one hour with monoclonal antibody mAb18B7 and then combined with secondary antibody (Alexa Fluror anti-mouse IgG 2 mg ml−1, Invitrogen). Cells were visualized on a Zeiss LSM inverted confocal microscope with the Argon/2 488 laser at 100×magnification.
Mating and Cell Fusion Assays
In a mating assay, strains of opposite mating type were mixed and co-cultured on V8 agar medium at 25°C in the dark. Mating filaments and basidiospore formation were examined and recorded by photography using a Zeiss Axio Imager A1 fluorescent microscope equipped with an AxioCam MRM digital camera after two weeks. Also, a cell fusion assay was performed [100]. 107 cells ml−1 of each mutant containing NatR or NeoR markers (DF001 and DF002) were mixed together in an equal volume and 5 µl of the mixture was spotted onto V8 medium and incubated for 24 h at 25°C in the dark. Scraped cells from the V8 medium, were resuspended in 1 ml dH2O, and then 200 µl of cell suspension were spread onto YPD medium containing nourseothricin and neomycin. The numbers of colonies on each plate were counted after 4 days of incubation at room temperature. TOC4 (Neor wild-type strain) and DF010 (Natr wild-type strain) were used as control strains.
Macrophage Killing Assays
We assessed ubp5Δ mutant and its complemented strains for survival within macrophages. Yeast strains were incubated overnight at 30°C. J774A.1 macrophages at a concentration of 106 per well were activated by gamma interferon-γ and lipopolysaccharide, then inoculated with yeasts for the macrophage-yeast survival assay [94]. Briefly, activated macrophages were coincubated with 106 yeast cells of each strain for 2 h at 37°C in 5% CO2 to allow phagocytosis. Prior to co-incubation, cells of each yeast strain were incubated with the monoclonal antibody mAb18B7 for one hour. Then extracellular yeasts were washed away three times with 1×PBS and the monolayers were incubated in DMEM. For the macrophage-yeast killing assay, washed monolayer cells containing yeast and macrophage cells were co-incubated overnight (24 hr). Macrophages were then lysed with 0.5% SDS, and lysates were diluted and plated on YPD to count numbers of viable yeasts present following incubation at 30°C for 3 days.
Galleria Mellonella Virulence Assay
G. mellonella caterpillars (330±25 mg in body weight) in the final instar larval stage were employed in our assays. Sixteen caterpillars of the required weight were randomly chosen in each group. A 10 ul Hamilton syringe was used to inject 10 ul aliquots of the yeast inoculum (5×105 cells) into the hemocoel of each caterpillar via the last left proleg as described previously [52], and these caterpillars were incubated in plastic containers at room temperature. Caterpillars were considered dead when they displayed no movement in response to touch. The number of dead caterpillars was recorded daily. Each assay was performed in duplicate.
Murine Virulence Assay
Virulence of the ubp5Δ mutant strain was assessed in the inhalational mouse model according to an established protocol [9]. A/Jcr mice (10 mice for each C. neoformans strain) were infected intranasally with 105 CFU of either WT (H99), mutant (ubp5Δ), or reconstituted (ubp5Δ+ UBP5) strains of C. neoformans in a volume of 50 µl sterile PBS. The mice were monitored closely for signs of worsening infection and euthanized at predetermined endpoints (weight loss of ≥15%, neurological symptoms, and an inability to access food/water).
Statistics
Data from the survival assays in both G. mellonella and murine infection models were plotted using Kaplan-Meier survival curves and analyzed by the log rank test (SPSS 15.0). All the other statistical data were calculated and compared using (Student t tests). P<0.05 were considered statistically significant.
Ethics Statement
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Institutional Animal Care and Use Committee of Duke University (Protocol Number: A266-08-10). Infections were carried out under isofluorane anesthesia, and all efforts were made to minimize suffering.
Supporting Information
Growth rate measurement for the ubp5Δ mutant. WT, ubp5Δ and ubp5Δ+UBP5 strains were grown overnight at 30°C in liquid YPD medium. Cell numbers were counted by hemacytometer. 106 CFU from each culture were transferred to 30 ml fresh YPD medium in flasks and incubated at 30°C. OD600 was measured for each culture at four hour intervals. Mutant ubp5Δ showed slower growth than H99 at 30°C.
(TIF)
Urease assay. Different DUB mutants grew on Christensen’s medium at 30°C for the detection of urease production. ure1Δ mutant strain was used as a negative control.
(TIF)
UBP5 deletion lead to increased expression of UBI4 in C. neoformans . Expression of polyubiquitin gene UBI4 at (A) 30°C and (B) 37°C. UBI4 was up-regulated in ubp5Δ mutant while the reconstituted strain showed similar expression level like WT strain. And high temperature exposure reinforced the up-regulation of UBI4 expression in ubp5Δ strain.
(TIF)
Primers used in this study. Primers were designed using the Primer3 software package (http://frodo.wi.mit.edu/primer3/) for generating gene-deletion cassettes, gene-reconstitution cassettes, Southern hybridization probes, or real-time PCR products.
(PDF)
Ubiquitin-proteasome genes identified by in vivo transcription profiling. Transcription profiles of in vivo C. neoformans cells from human or rabbit infections was compared to profiles from C. neoformans cells grown ex vivo in either pooled human cerebrospinal fluid or serum to identify genes that are differentially expressed in the host.
(PDF)
Acknowledgments
We thank Connie Nichols and Andrew Alspaugh for thoughtful discussions of the data and its implications.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was funded by National Institutes of Health PHS grants AI73896 and AI28388 (JRP), and also supported by Shanghai Science and Technology Committee grant 10dz2220100 (W-QL). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, et al. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS. 2009;23:525–530. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- 2.Chayakulkeeree M, Perfect JR. Cryptococcosis. Infect Dis Clin North Am 20: 507–544, v-vi. 2006. [DOI] [PubMed]
- 3.Kidd SE, Hagen F, Tscharke RL, Huynh M, Bartlett KH, et al. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). Proc Natl Acad Sci U S A. 2004;101:17258–17263. doi: 10.1073/pnas.0402981101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown SM, Campbell LT, Lodge JK. Cryptococcus neoformans, a fungus under stress. Curr Opin Microbiol. 2007;10:320–325. doi: 10.1016/j.mib.2007.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown AJ, Haynes K, Quinn J. Nitrosative and oxidative stress responses in fungal pathogenicity. Curr Opin Microbiol. 2009;12:384–391. doi: 10.1016/j.mib.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang YC, Kwon-Chung KJ. Complementation of a capsule-deficient mutation of Cryptococcus neoformans restores its virulence. Mol Cell Biol. 1994;14:4912–4919. doi: 10.1128/mcb.14.7.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu X, Williamson PR. Role of laccase in the biology and virulence of Cryptococcus neoformans. FEMS Yeast Res. 2004;5:1–10. doi: 10.1016/j.femsyr.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Perfect JR. Cryptococcus neoformans: the yeast that likes it hot. FEMS Yeast Res. 2006;6:463–468. doi: 10.1111/j.1567-1364.2006.00051.x. [DOI] [PubMed] [Google Scholar]
- 9.Cox GM, Mukherjee J, Cole GT, Casadevall A, Perfect JR. Urease as a virulence factor in experimental cryptococcosis. Infect Immun. 2000;68:443–448. doi: 10.1128/iai.68.2.443-448.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox GM, McDade HC, Chen SC, Tucker SC, Gottfredsson M, et al. Extracellular phospholipase activity is a virulence factor for Cryptococcus neoformans. Mol Microbiol. 2001;39:166–175. doi: 10.1046/j.1365-2958.2001.02236.x. [DOI] [PubMed] [Google Scholar]
- 11.Wang P, Heitman J. Signal transduction cascades regulating mating, filamentation, and virulence in Cryptococcus neoformans. Curr Opin Microbiol. 1999;2:358–362. doi: 10.1016/S1369-5274(99)80063-0. [DOI] [PubMed] [Google Scholar]
- 12.Pukkila-Worley R, Gerrald QD, Kraus PR, Boily MJ, Davis MJ, et al. Transcriptional network of multiple capsule and melanin genes governed by the Cryptococcus neoformans cyclic AMP cascade. Eukaryot Cell. 2005;4:190–201. doi: 10.1128/EC.4.1.190-201.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kozubowski L, Lee SC, Heitman J. Signalling pathways in the pathogenesis of Cryptococcus. Cell Microbiol. 2009;11:370–380. doi: 10.1111/j.1462-5822.2008.01273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hull CM, Heitman J. Genetics of Cryptococcus neoformans. Annu Rev Genet. 2002;36:557–615. doi: 10.1146/annurev.genet.36.052402.152652. [DOI] [PubMed] [Google Scholar]
- 15.Idnurm A, Bahn YS, Nielsen K, Lin X, Fraser JA, et al. Deciphering the model pathogenic fungus Cryptococcus neoformans. Nat Rev Microbiol. 2005;3:753–764. doi: 10.1038/nrmicro1245. [DOI] [PubMed] [Google Scholar]
- 16.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 17.Finley D, Ciechanover A, Varshavsky A. Ubiquitin as a central cellular regulator. Cell 116: S29–32, 2 p following S32. 2004. [DOI] [PubMed]
- 18.Wilkinson KD. Ubiquitin: a Nobel protein. Cell. 2004;119:741–745. doi: 10.1016/j.cell.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 19.Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 20.Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 21.Kouranti I, McLean JR, Feoktistova A, Liang P, Johnson AE, et al. A global census of fission yeast deubiquitinating enzyme localization and interaction networks reveals distinct compartmentalization profiles and overlapping functions in endocytosis and polarity. LID - e1000471 [pii]. PLoS Biol 8. 2010. [DOI] [PMC free article] [PubMed]
- 22.Amerik AY, Li SJ, Hochstrasser M. Analysis of the deubiquitinating enzymes of the yeast Saccharomyces cerevisiae. Biol Chem. 2000;381:981–992. doi: 10.1515/BC.2000.121. [DOI] [PubMed] [Google Scholar]
- 23.Komander D, Clague MJ, Urbe S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 24.Kahana A. The deubiquitinating enzyme Dot4p is involved in regulating nutrient uptake. Biochem Biophys Res Commun. 2001;282:916–920. doi: 10.1006/bbrc.2001.4669. [DOI] [PubMed] [Google Scholar]
- 25.Enyenihi AH, Saunders WS. Large-scale functional genomic analysis of sporulation and meiosis in Saccharomyces cerevisiae. Genetics. 2003;163:47–54. doi: 10.1093/genetics/163.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dudley AM, Janse DM, Tanay A, Shamir R, Church GM. A global view of pleiotropy and phenotypically derived gene function in yeast. Mol Syst Biol. 2005;1:2005.0001. doi: 10.1038/msb4100004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weiss P, Huppert S, Kolling R. ESCRT-III protein Snf7 mediates high-level expression of the SUC2 gene via the Rim101 pathway. Eukaryot Cell. 2008;7:1888–1894. doi: 10.1128/EC.00194-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Auesukaree C, Damnernsawad A, Kruatrachue M, Pokethitiyook P, Boonchird C, et al. Genome-wide identification of genes involved in tolerance to various environmental stresses in Saccharomyces cerevisiae. J Appl Genet. 2009;50:301–310. doi: 10.1007/BF03195688. [DOI] [PubMed] [Google Scholar]
- 29.Eisele F, Braun B, Pfirrmann T, Wolf DH. Mutants of the deubiquitinating enzyme Ubp14 decipher pathway diversity of ubiquitin-proteasome linked protein degradation. Biochem Biophys Res Commun. 2006;350:329–333. doi: 10.1016/j.bbrc.2006.09.041. [DOI] [PubMed] [Google Scholar]
- 30.Alamgir M, Erukova V, Jessulat M, Azizi A, Golshani A. Chemical-genetic profile analysis of five inhibitory compounds in yeast. BMC Chem Biol. 2010;10:6. doi: 10.1186/1472-6769-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshikawa K, Tanaka T, Ida Y, Furusawa C, Hirasawa T, et al. Comprehensive phenotypic analysis of single-gene deletion and overexpression strains of Saccharomyces cerevisiae. Yeast. 2011;28:349–361. doi: 10.1002/yea.1843. [DOI] [PubMed] [Google Scholar]
- 32.Liu OW, Chun CD, Chow ED, Chen C, Madhani HD, et al. Systematic genetic analysis of virulence in the human fungal pathogen Cryptococcus neoformans. Cell. 2008;135:174–188. doi: 10.1016/j.cell.2008.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ko YJ, Yu YM, Kim GB, Lee GW, Maeng PJ, et al. Remodeling of global transcription patterns of Cryptococcus neoformans genes mediated by the stress-activated HOG signaling pathways. Eukaryot Cell. 2009;8:1197–1217. doi: 10.1128/EC.00120-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilkinson KD. Regulation of ubiquitin-dependent processes by deubiquitinating enzymes. FASEB J. 1997;11:1245–1256. doi: 10.1096/fasebj.11.14.9409543. [DOI] [PubMed] [Google Scholar]
- 35.Petzold EW, Himmelreich U, Mylonakis E, Rude T, Toffaletti D, et al. Characterization and regulation of the trehalose synthesis pathway and its importance in the pathogenicity of Cryptococcus neoformans. Infect Immun. 2006;74:5877–5887. doi: 10.1128/IAI.00624-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoneda A, Doering TL. A eukaryotic capsular polysaccharide is synthesized intracellularly and secreted via exocytosis. Mol Biol Cell. 2006;17:5131–5140. doi: 10.1091/mbc.E06-08-0701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reese AJ, Doering TL. Cell wall alpha-1,3-glucan is required to anchor the Cryptococcus neoformans capsule. Mol Microbiol. 2003;50:1401–1409. doi: 10.1046/j.1365-2958.2003.03780.x. [DOI] [PubMed] [Google Scholar]
- 38.McClelland EE, Bernhardt P, Casadevall A. Estimating the relative contributions of virulence factors for pathogenic microbes. Infect Immun. 2006;74:1500–1504. doi: 10.1128/IAI.74.3.1500-1504.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walton FJ, Idnurm A, Heitman J. Novel gene functions required for melanization of the human pathogen Cryptococcus neoformans. Mol Microbiol. 2005;57:1381–1396. doi: 10.1111/j.1365-2958.2005.04779.x. [DOI] [PubMed] [Google Scholar]
- 40.Jiang N, Sun N, Xiao D, Pan J, Wang Y, et al. A copper-responsive factor gene CUF1 is required for copper induction of laccase in Cryptococcus neoformans. FEMS Microbiol Lett. 2009;296:84–90. doi: 10.1111/j.1574-6968.2009.01619.x. [DOI] [PubMed] [Google Scholar]
- 41.Liu J, Sitaram A, Burd CG. Regulation of copper-dependent endocytosis and vacuolar degradation of the yeast copper transporter, Ctr1p, by the Rsp5 ubiquitin ligase. Traffic. 2007;8:1375–1384. doi: 10.1111/j.1600-0854.2007.00616.x. [DOI] [PubMed] [Google Scholar]
- 42.Brady GF, Galban S, Liu X, Basrur V, Gitlin JD, et al. Regulation of the copper chaperone CCS by XIAP-mediated ubiquitination. Mol Cell Biol. 2010;30:1923–1936. doi: 10.1128/MCB.00900-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olszewski MA, Noverr MC, Chen GH, Toews GB, Cox GM, et al. Urease expression by Cryptococcus neoformans promotes microvascular sequestration, thereby enhancing central nervous system invasion. Am J Pathol. 2004;164:1761–1771. doi: 10.1016/S0002-9440(10)63734-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Osterholzer JJ, Surana R, Milam JE, Montano GT, Chen GH, et al. Cryptococcal urease promotes the accumulation of immature dendritic cells and a non-protective T2 immune response within the lung. Am J Pathol. 2009;174:932–943. doi: 10.2353/ajpath.2009.080673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davidson RC, Nichols CB, Cox GM, Perfect JR, Heitman J. A MAP kinase cascade composed of cell type specific and non-specific elements controls mating and differentiation of the fungal pathogen Cryptococcus neoformans. Mol Microbiol. 2003;49:469–485. doi: 10.1046/j.1365-2958.2003.03563.x. [DOI] [PubMed] [Google Scholar]
- 46.Shen WC, Davidson RC, Cox GM, Heitman J. Pheromones stimulate mating and differentiation via paracrine and autocrine signaling in Cryptococcus neoformans. Eukaryot Cell. 2002;1:366–377. doi: 10.1128/EC.1.3.366-377.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feldmesser M, Kress Y, Novikoff P, Casadevall A. Cryptococcus neoformans is a facultative intracellular pathogen in murine pulmonary infection. Infect Immun. 2000;68:4225–4237. doi: 10.1128/iai.68.7.4225-4237.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim KS. Mechanisms of microbial traversal of the blood-brain barrier. Nat Rev Microbiol. 2008;6:625–634. doi: 10.1038/nrmicro1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y, Aisen P, Casadevall A. Cryptococcus neoformans melanin and virulence: mechanism of action. Infect Immun. 1995;63:3131–3136. doi: 10.1128/iai.63.8.3131-3136.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cox GM, Harrison TS, McDade HC, Taborda CP, Heinrich G, et al. Superoxide dismutase influences the virulence of Cryptococcus neoformans by affecting growth within macrophages. Infect Immun. 2003;71:173–180. doi: 10.1128/IAI.71.1.173-180.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Jesus-Berrios M, Liu L, Nussbaum JC, Cox GM, Stamler JS, et al. Enzymes that counteract nitrosative stress promote fungal virulence. Curr Biol. 2003;13:1963–1968. doi: 10.1016/j.cub.2003.10.029. [DOI] [PubMed] [Google Scholar]
- 52.Mylonakis E, Moreno R, El KJB, Idnurm A, Heitman J, et al. Galleria mellonella as a model system to study Cryptococcus neoformans pathogenesis. Infect Immun. 2005;73:3842–3850. doi: 10.1128/IAI.73.7.3842-3850.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Velagapudi R, Hsueh YP, Geunes-Boyer S, Wright JR, Heitman J. Spores as infectious propagules of Cryptococcus neoformans. Infect Immun. 2009;77:4345–4355. doi: 10.1128/IAI.00542-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kozubowski L, Heitman J. Septins enforce morphogenetic events during sexual reproduction and contribute to virulence of Cryptococcus neoformans. Mol Microbiol. 2010;75:658–675. doi: 10.1111/j.1365-2958.2009.06983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Narasipura SD, Chaturvedi V, Chaturvedi S. Characterization of Cryptococcus neoformans variety gattii SOD2 reveals distinct roles of the two superoxide dismutases in fungal biology and virulence. Mol Microbiol. 2005;55:1782–1800. doi: 10.1111/j.1365-2958.2005.04503.x. [DOI] [PubMed] [Google Scholar]
- 56.Semighini CP, Averette AF, Perfect JR, Heitman J. Deletion of Cryptococcus neoformans AIF Ortholog Promotes Chromosome Aneuploidy and Fluconazole-Resistance in a Metacaspase-Independent Manner. PLoS Pathog. 2011;7:e1002364. doi: 10.1371/journal.ppat.1002364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, et al. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 58.Boase NA, Lockington RA, Adams JR, Rodbourn L, Kelly JM. Molecular characterization and analysis of the acrB gene of Aspergillus nidulans: a gene identified by genetic interaction as a component of the regulatory network that includes the CreB deubiquitination enzyme. Genetics. 2003;164:95–104. doi: 10.1093/genetics/164.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang J, Schneider C, Ottmers L, Rodriguez R, Day A, et al. Genomic scale mutant hunt identifies cell size homeostasis genes in S. cerevisiae. Curr Biol. 2002;12:1992–2001. doi: 10.1016/s0960-9822(02)01305-2. [DOI] [PubMed] [Google Scholar]
- 60.Bozza WP, Zhuang Z. Biochemical characterization of a multidomain deubiquitinating enzyme ubp15 and the regulatory role of its terminal domains. Biochemistry. 2011;50:6423–6432. doi: 10.1021/bi200529z. [DOI] [PubMed] [Google Scholar]
- 61.Goranov AI, Cook M, Ricicova M, Ben-Ari G, Gonzalez C, et al. The rate of cell growth is governed by cell cycle stage. Genes Dev. 2009;23:1408–1422. doi: 10.1101/gad.1777309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ellis DH, Pfeiffer TJ. Ecology, life cycle, and infectious propagule of Cryptococcus neoformans. Lancet. 1990;336:923–925. doi: 10.1016/0140-6736(90)92283-n. [DOI] [PubMed] [Google Scholar]
- 63.Lin X. Cryptococcus neoformans: morphogenesis, infection, and evolution. Infect Genet Evol. 2009;9:401–416. doi: 10.1016/j.meegid.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 64.Liu TB, Wang Y, Stukes S, Chen Q, Casadevall A, et al. The F-box protein Fbp1 regulates sexual reproduction and virulence in Cryptococcus neoformans. Eukaryot Cell. 2011. [DOI] [PMC free article] [PubMed]
- 65.Finley D, Ozkaynak E, Varshavsky A. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell. 1987;48:1035–1046. doi: 10.1016/0092-8674(87)90711-2. [DOI] [PubMed] [Google Scholar]
- 66.Johnson PR, Swanson R, Rakhilina L, Hochstrasser M. Degradation signal masking by heterodimerization of MATalpha2 and MATa1 blocks their mutual destruction by the ubiquitin-proteasome pathway. Cell. 1998;94:217–227. doi: 10.1016/s0092-8674(00)81421-x. [DOI] [PubMed] [Google Scholar]
- 67.Esch RK, Errede B. Pheromone induction promotes Ste11 degradation through a MAPK feedback and ubiquitin-dependent mechanism. Proc Natl Acad Sci U S A. 2002;99:9160–9165. doi: 10.1073/pnas.142034399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Laney JD, Hochstrasser M. Ubiquitin-dependent degradation of the yeast Mat(alpha)2 repressor enables a switch in developmental state. Genes Dev. 2003;17:2259–2270. doi: 10.1101/gad.1115703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gancedo C, Flores CL. The importance of a functional trehalose biosynthetic pathway for the life of yeasts and fungi. FEMS Yeast Res. 2004;4:351–359. doi: 10.1016/S1567-1356(03)00222-8. [DOI] [PubMed] [Google Scholar]
- 70.Nichols CB, Perfect ZH, Alspaugh JA. A Ras1-Cdc24 signal transduction pathway mediates thermotolerance in the fungal pathogen Cryptococcus neoformans. Mol Microbiol. 2007;63:1118–1130. doi: 10.1111/j.1365-2958.2006.05566.x. [DOI] [PubMed] [Google Scholar]
- 71.Alspaugh JA, Cavallo LM, Perfect JR, Heitman J. RAS1 regulates filamentation, mating and growth at high temperature of Cryptococcus neoformans. Mol Microbiol. 2000;36:352–365. doi: 10.1046/j.1365-2958.2000.01852.x. [DOI] [PubMed] [Google Scholar]
- 72.Ballou ER, Nichols CB, Miglia KJ, Kozubowski L, Alspaugh JA. Two CDC42 paralogues modulate Cryptococcus neoformans thermotolerance and morphogenesis under host physiological conditions. Mol Microbiol. 2010;75:763–780. doi: 10.1111/j.1365-2958.2009.07019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vandenbroucke K, Robbens S, Vandepoele K, Inze D, Van de Peer Y, et al. Hydrogen peroxide-induced gene expression across kingdoms: a comparative analysis. Mol Biol Evol. 2008;25:507–516. doi: 10.1093/molbev/msm276. [DOI] [PubMed] [Google Scholar]
- 74.Treger JM, Heichman KA, McEntee K. Expression of the yeast UB14 gene increases in response to DNA-damaging agents and in meiosis. Mol Cell Biol. 1988;8:1132–1136. doi: 10.1128/mcb.8.3.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cheng L, Watt R, Piper PW. Polyubiquitin gene expression contributes to oxidative stress resistance in respiratory yeast (Saccharomyces cerevisiae). Mol Gen Genet. 1994;243:358–362. doi: 10.1007/BF00301072. [DOI] [PubMed] [Google Scholar]
- 76.Spitzer ED, Spitzer SG. Structure of the ubiquitin-encoding genes of Cryptococcus neoformans. Gene. 1995;161:113–117. doi: 10.1016/0378-1119(95)00231-t. [DOI] [PubMed] [Google Scholar]
- 77.Bahn YS. Master and commander in fungal pathogens: the two-component system and the HOG signaling pathway. Eukaryot Cell. 2008;7:2017–2036. doi: 10.1128/EC.00323-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sato N, Kawahara H, Toh-e A, Maeda T. Phosphorelay-regulated degradation of the yeast Ssk1p response regulator by the ubiquitin-proteasome system. Mol Cell Biol. 2003;23:6662–6671. doi: 10.1128/MCB.23.18.6662-6671.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kishi T, Ikeda A, Nagao R, Koyama N. The SCFCdc4 ubiquitin ligase regulates calcineurin signaling through degradation of phosphorylated Rcn1, an inhibitor of calcineurin. Proc Natl Acad Sci U S A. 2007;104:17418–17423. doi: 10.1073/pnas.0704951104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cappell SD, Baker R, Skowyra D, Dohlman HG. Systematic analysis of essential genes reveals important regulators of G protein signaling. Mol Cell. 2010;38:746–757. doi: 10.1016/j.molcel.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ding F, Yin Z, Wang HR. Ubiquitination in Rho signaling. Curr Top Med Chem. 2011. [DOI] [PubMed]
- 82.Rhodes JC, Polacheck I, Kwon-Chung KJ. Phenoloxidase activity and virulence in isogenic strains of Cryptococcus neoformans. Infect Immun. 1982;36:1175–1184. doi: 10.1128/iai.36.3.1175-1184.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen SC, Muller M, Zhou JZ, Wright LC, Sorrell TC. Phospholipase activity in Cryptococcus neoformans: a new virulence factor. J Infect Dis. 1997;175:414–420. doi: 10.1093/infdis/175.2.414. [DOI] [PubMed] [Google Scholar]
- 84.Chen SC, Wright LC, Santangelo RT, Muller M, Moran VR, et al. Identification of extracellular phospholipase B, lysophospholipase, and acyltransferase produced by Cryptococcus neoformans. Infect Immun. 1997;65:405–411. doi: 10.1128/iai.65.2.405-411.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Doering TL. How sweet it is! Cell wall biogenesis and polysaccharide capsule formation in Cryptococcus neoformans. Annu Rev Microbiol. 2009;63:223–247. doi: 10.1146/annurev.micro.62.081307.162753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Reese AJ, Yoneda A, Breger JA, Beauvais A, Liu H, et al. Loss of cell wall alpha(1–3) glucan affects Cryptococcus neoformans from ultrastructure to virulence. Mol Microbiol. 2007;63:1385–1398. doi: 10.1111/j.1365-2958.2006.05551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rodrigues ML, Alvarez M, Fonseca FL, Casadevall A. Binding of the wheat germ lectin to Cryptococcus neoformans suggests an association of chitinlike structures with yeast budding and capsular glucuronoxylomannan. Eukaryot Cell. 2008;7:602–609. doi: 10.1128/EC.00307-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.O’Meara TR, Norton D, Price MS, Hay C, Clements MF, et al. Interaction of Cryptococcus neoformans Rim101 and protein kinase A regulates capsule. PLoS Pathog. 2010;6:e1000776. doi: 10.1371/journal.ppat.1000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Huffnagle GB, Chen GH, Curtis JL, McDonald RA, Strieter RM, et al. Down-regulation of the afferent phase of T cell-mediated pulmonary inflammation and immunity by a high melanin-producing strain of Cryptococcus neoformans. J Immunol. 1995;155:3507–3516. [PubMed] [Google Scholar]
- 90.Steenbergen JN, Shuman HA, A C. Cryptococcus neoformans interactions with amoebae suggest an explanation for its virulence and intracellular pathogenic strategy in macrophages. 2001;98:15245–15250. doi: 10.1073/pnas.261418798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nicholson B, Marblestone JG, Butt TR, Mattern MR. Deubiquitinating enzymes as novel anticancer targets. Future Oncol. 2007;3:191–199. doi: 10.2217/14796694.3.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cox GM, Mukherjee J, Cole GT, Casadevall A, Perfect JR. Urease as a virulence factor in experimental cryptococcosis. Infect Immun. 2000;68:443–448. doi: 10.1128/iai.68.2.443-448.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Price MS, Nichols CB, Alspaugh JA. The Cryptococcus neoformans Rho-GDP dissociation inhibitor mediates intracellular survival and virulence. Infect Immun. 2008;76:5729–5737. doi: 10.1128/IAI.00896-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lee A, Toffaletti DL, Tenor J, Soderblom EJ, Thompson JW, et al. Survival defects of Cryptococcus neoformans mutants exposed to human cerebrospinal fluid result in attenuated virulence in an experimental model of meningitis. Infect Immun. 2010;78:4213–4225. doi: 10.1128/IAI.00551-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lee A, Toffaletti DL, Tenor J, Soderblom EJ, Thompson JW, et al. Survival defects of Cryptococcus neoformans mutants exposed to human cerebrospinal fluid result in attenuated virulence in an experimental model of meningitis. Infect Immun. 2010;78:4213–4225. doi: 10.1128/IAI.00551-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Price MS, Betancourt-Quiroz M, Price JL, Toffaletti DL, Vora H, et al. Cryptococcus neoformans requires a functional glycolytic pathway for disease but not persistence in the host. MBio. 2011;2:e00103–00111. doi: 10.1128/mBio.00103-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Davidson RC, Blankenship JR, Kraus PR, de Jesus Berrios M, Hull CM, et al. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology. 2002;148:2607–2615. doi: 10.1099/00221287-148-8-2607. [DOI] [PubMed] [Google Scholar]
- 98.Toffaletti DL, Rude TH, Johnston SA, Durack DT, Perfect JR. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J Bacteriol. 1993;175:1405–1411. doi: 10.1128/jb.175.5.1405-1411.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Varma A, Kwon-Chung KJ. Characterization of the glyceraldehyde-3-phosphate dehydrogenase gene [correction of glyceraldehyde-3-phosphate gene] and the use of its promoter for heterologous expression in Cryptococcus neoformans, a human pathogen. Gene. 1999;232:155–163. doi: 10.1016/s0378-1119(99)00132-8. [DOI] [PubMed] [Google Scholar]
- 100.Bahn YS, Kojima K, Cox GM, Heitman J. A unique fungal two-component system regulates stress responses, drug sensitivity, sexual development, and virulence of Cryptococcus neoformans. Mol Biol Cell. 2006;17:3122–3135. doi: 10.1091/mbc.E06-02-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nielsen K, Cox GM, Wang P, Toffaletti DL, Perfect JR, et al. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and alpha isolates. Infect Immun. 2003;71:4831–4841. doi: 10.1128/IAI.71.9.4831-4841.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Growth rate measurement for the ubp5Δ mutant. WT, ubp5Δ and ubp5Δ+UBP5 strains were grown overnight at 30°C in liquid YPD medium. Cell numbers were counted by hemacytometer. 106 CFU from each culture were transferred to 30 ml fresh YPD medium in flasks and incubated at 30°C. OD600 was measured for each culture at four hour intervals. Mutant ubp5Δ showed slower growth than H99 at 30°C.
(TIF)
Urease assay. Different DUB mutants grew on Christensen’s medium at 30°C for the detection of urease production. ure1Δ mutant strain was used as a negative control.
(TIF)
UBP5 deletion lead to increased expression of UBI4 in C. neoformans . Expression of polyubiquitin gene UBI4 at (A) 30°C and (B) 37°C. UBI4 was up-regulated in ubp5Δ mutant while the reconstituted strain showed similar expression level like WT strain. And high temperature exposure reinforced the up-regulation of UBI4 expression in ubp5Δ strain.
(TIF)
Primers used in this study. Primers were designed using the Primer3 software package (http://frodo.wi.mit.edu/primer3/) for generating gene-deletion cassettes, gene-reconstitution cassettes, Southern hybridization probes, or real-time PCR products.
(PDF)
Ubiquitin-proteasome genes identified by in vivo transcription profiling. Transcription profiles of in vivo C. neoformans cells from human or rabbit infections was compared to profiles from C. neoformans cells grown ex vivo in either pooled human cerebrospinal fluid or serum to identify genes that are differentially expressed in the host.
(PDF)