Abstract
Deficiency of fatty acid translocase Cd36 has been shown to have a major role in the pathogenesis of metabolic syndrome in the spontaneously hypertensive rat (SHR). We have tested the hypothesis that the effects of Cd36 mutation on the features of metabolic syndrome are contextually dependent on genomic background. We have derived two new congenic strains by introgression of limited chromosome 4 regions of SHR origin, both including the defective Cd36 gene, into the genetic background of a highly inbred model of insulin resistance and dyslipidemia, polydactylous (PD) rat strain. We subjected standard diet-fed adult males of PD and the congenic PD.SHR4 strains to metabolic, morphometric and transcriptomic profiling. We observed significantly improved glucose tolerance and lower fasting insulin levels in PD.SHR4 congenics than in PD. One of the PD.SHR4 strains showed lower triglyceride concentrations across major lipoprotein fractions combined with higher levels of low-density lipoprotein cholesterol compared with the PD progenitor. The hepatic transcriptome assessment revealed a network of genes differentially expressed between PD and PD.SHR4 with significant enrichment by members of the circadian rhythmicity pathway (Arntl (Bmal1), Clock, Nfil3, Per2 and Per3). In summary, the introduction of the chromosome 4 region of SHR origin including defective Cd36 into the PD genetic background resulted in disconnected shifts of metabolic profile along with distinct changes in hepatic transcriptome. The synthesis of the current results with those obtained in other Cd36-deficient strains indicates that the eventual metabolic effect of a deleterious mutation such as that of SHR-derived Cd36 is not absolute, but rather a function of complex interactions between environmental and genomic background, upon which it operates.
Keywords: functional genomics, metabolic syndrome, animal model, transcriptomics
Introduction
Metabolic syndrome is a complex human condition with rising prevalence worldwide. Recently, several previously used definitions have been merged into a unified definition of the metabolic syndrome (Alberti et al., 2009). This definition lists dyslipidemia, obesity, hypertension and fasting glucose levels among the syndrome's major components. In spite of clear evidence of the ecogenomic (genome × environment interactive) nature of the pathogenesis of the syndrome and its individual aspects, the identification of causal genetic variants remains elusive even in the era of large, genome-wide association studies (Kraja et al., 2011). To date, one of the few successfully and positionally cloned culprits linked to many facets of the metabolic syndrome is a gene coding for fatty acid translocase Cd36, mutation of which has been originally found in the spontaneously hypertensive rat (SHR) (Aitman et al., 1997, 1999; Pravenec et al., 2001). Interestingly, the complex nature of mutation of Cd36 (a chimeric gene owing to unequal recombination between the Cd36 gene and one of its pseudogenes) in SHR (Glazier et al., 2002) does not result in complete lack of expression, but rather in severe, tissue-specific downregulation of the gene (Bonen et al., 2009). Although rare, complete deficiencies of Cd36 were reported in humans (Yamamoto et al., 1994; Kashiwagi et al., 1996) and a murine model featuring complete knockout of Cd36 is available (Febbraio et al., 1999), models carrying the ‘incomplete' type of mutation may relevantly represent the more subtle functional consequences of common polymorphisms in human Cd36 gene. Actually, several single-nucleotide polymorphisms in the human Cd36 gene were recently associated with lipid levels, metabolic syndrome (Love-Gregory et al., 2008; Noel et al., 2010) or obesity (Bokor et al., 2010), although the latter has not been unanimously confirmed (Choquet et al., 2010).
We have previously shown that introgression of rat chromosome 4 segment including the mutant Cd36 gene from SHR into the Brown Norway genome results in deterioration of insulin sensitivity and increase in triacylglycerol and free fatty acid levels in the BN.SHR4 congenic strain (Seda et al., 2002). Furthermore, we have found this limited portion of genome to be crucial for several pharmacogenetic interactions involving the insulin sensitizer rosiglitazone (Seda et al., 2003a, 2008) and glucocorticoid dexamethasone (Krupkova et al., 2010). In SHR, Cd36 was established as a key determinant of the insulin-sensitizing actions of thiazolidinediones using SHR transgenic and congenic strains expressing wild-type Cd36 (Qi et al., 2002). In this study, we have tested the metabolic and transcriptomic impact of the presence of defective Cd36 within the genomic background of a highly inbred model of metabolic syndrome, the polydactylous rat (Kren, 1975; Sedova et al., 2000). The choice was driven by our previous differential linkage study conducted in PD × BN.SHR4 cross, in which we mapped several suggestive and significant quantitative-trait loci of metabolic syndrome-related traits in the region of Cd36/Fat gene (Seda et al., 2003b).
Materials and methods
Rat strains
The polydactylous rat strain (PD/Cub, PD hereafter, Rat Genome Database (Dwinell et al., 2009) (RGD) ID no. 728161) is a highly inbred strain (F>90) showing metabolic syndrome attributes (Sedova et al., 2000; Seda et al., 2005), kept since 1969 at the Institute of Biology and Medical Genetics, First Faculty of Medicine, Charles University in Prague (Kren, 1975). In this study, we have established two new congenic strains combining the genomic information of two established genetic models of metabolic syndrome, PD and the spontaneously hypertensive rat (SHR/OlaIpcv, SHR hereafter, RGD ID no. 631848). Such a combination was achieved by introgression of the chromosome 4 segment of SHR origin into the PD genetic background, using marker-assisted backcross breeding. After verifying the congenicity of both the new strains by a whole-genomic marker scan, we precisely defined the extent of SHR-derived regions by genotyping 54 polymorphic microsatellite markers (Table 1).
Table 1. Definition of the differential segments in the new PD.SHR4 congenic strains.
| Marker | Mbp | PD.SHR4a | PD.SHR4b | Selected genes differing in origin between PD.SHR4a and PD.SHR4b |
|---|---|---|---|---|
| D4Rat248 | 0.28 | PD | SHR | |
| D4Arb14 | 0.45 | PD | SHR | |
| Il6 | 0.46 | PD | SHR | Il6 |
| D4Rat117 | 1.18 | PD | SHR | Dnajb6 |
| Lmbr1_i4 | 1.25 | PD | SHR | Ube3c |
| Lmbr_i5_120-816 | 1.26 | PD | SHR | Lmbr |
| D4Mgh7 | 1.55 | PD | PD | |
| D4Rat147 | 2.45 | PD | SHR | Shh, En2, Insig1 |
| D4Rat139 | 3.04 | PD | SHR | Htr5a, Paxip1, Dpp6 |
| D4Arb13 | 3.17 | SHR | SHR | |
| D4Rat4 | 3.23 | SHR | SHR | |
| D4Rat142 | 4.82 | SHR | SHR | |
| D4Rat2 | 5.43 | SHR | SHR | |
| D4Bro1 | 6.12 | SHR | SHR | |
| D4Rat1 | 6.69 | SHR | SHR | |
| D4Rat136 | 8.48 | SHR | SHR | |
| D4Rat5 | 9.63 | SHR | SHR | |
| D4Rat148 | 9.68 | SHR | SHR | |
| D4Rat6 | 10.29 | SHR | SHR | |
| D4Rat7 | 10.67 | SHR | SHR | Cd36 |
| D4Rat221 | 14.38 | SHR | SHR | |
| D4Rat222 | 19.03 | SHR | SHR | |
| D4Rat125 | 19.46 | SHR | SHR | |
| D4Rat10 | 26.29 | PD | SHR | Grm3, Abcb1a, Abcb1b, Crot |
| D4Rat151 | 29.44 | PD | SHR | Cyp51, Pex1, Cdk6, Pon1 |
| D4Rat17 | 46.03 | PD | PD | |
| D4Rat16 | 49.33 | PD | PD | |
| D4Rat119 | 63.10 | PD | PD | |
| D4Rat102 | 66.49 | PD | PD | |
| D4Rat27 | 71.71 | PD | PD | |
| D4Rat28 | 75.34 | PD | PD | |
| D4Rat168 | 77.42 | PD | PD | |
| D4Mit24 | 78.32 | PD | PD | |
| D4Rat33 | 80.47 | PD | PD | |
| D4Rat34 | 85.25 | PD | PD | |
| D4Rat35 | 91.57 | PD | PD | |
| D4Rat172 | 95.16 | PD | PD | |
| D4Rat39 | 104.44 | PD | PD | |
| D4Rat175 | 106.72 | PD | PD | |
| D4Rat97 | 111.29 | PD | PD | |
| D4Rat176 | 116.55 | PD | PD | |
| D4Rat53 | 126.31 | PD | PD | |
| D4Rat80 | 132.50 | PD | PD | |
| D4Rat58 | 136.33 | PD | PD | |
| D4Rat196 | 143.99 | PD | PD | |
| D4Rat60 | 149.80 | PD | PD | |
| D4Rat59 | 150.28 | PD | PD | |
| D4Rat137 | 155.58 | PD | PD | |
| D4Rat203 | 161.67 | PD | PD | |
| D4Arb27 | 164.84 | PD | PD | |
| D4Rat68 | 172.40 | PD | PD | |
| D4Rat69 | 175.18 | PD | PD | |
| D4Rat140 | 182.98 | PD | PD | |
| D4Rat72 | 186.34 | PD | PD |
Abbreviations: PD, polydactylous; SHR, spontaneously hypertensive rat.
Experimental protocol
Our study was performed in conformity with the Animal Protection Law of the Czech Republic (311/1997), which is in compliance with the European Community Council recommendations for the use of laboratory animals 86/609/ECC and was approved by the Ethical committee of the First Faculty of Medicine. Adult rat males were held under temperature- and humidity-controlled conditions on a 12-h/12-h light–dark cycle. Throughout the study, the animals had free access to food (standard chow) and water. At the age of 4 months, adult males of both congenic strains and the parental strain PD (n=6–8 per strain) were subjected to oral glucose-tolerance test after overnight fasting and the blood samples for other metabolic measurements were drawn. Then the animals were killed and the total weight and the weight of heart, liver, kidneys, adrenals, epididymal and retroperitoneal fat pads were determined; liver tissue was snap frozen in liquid nitrogen for further analyses of gene expression.
DNA extraction, genotyping
PCR was used for genotyping markers polymorphic between progenitor strains. We tested the DNA of both congenic strains (PD.SHR4a, n=8; PD.SHR4b, n=6) and the progenitor strains PD/Cub and SHR. The rat DNA was isolated by a modified phenol-extraction method from tail-incision samples. Nucleotide sequences of primers were obtained from public databases (RGD, http://rgd.mcw.edu/, The Welcome Trust Centre for Human Genetics, http://www.well.ox.ac.uk/ or Whitehead Institute/MIT Center for Genome Research, http://www-genome.wi.mit.edu/). The PCR products were separated on polyacrylamide (7–10%) gels, detected in ultraviolet light after ethidium-bromide staining using Syngene G:Box.
Metabolic measurements
The oral glucose-tolerance test (OGTT) was performed after overnight fasting and the blood samples for the glycemia determination (Ascensia Elite Blood Glucose Meter; Bayer HealthCare, Mishawaka, IN, USA, validated by Institute of Clinical Biochemistry and Laboratory Diagnostics of the First Faculty of Medicine) were obtained from the tail vein at intervals of 0, 30, 60, 120 and 180 min after intragastric glucose administration to conscious rats (3 g kg−1 body weight, 30% aqueous solution). The lipid profile (cholesterol and triglyceride blood concentration in 20 lipoprotein fractions, glycerol level and chylomicron, very-low-density lipoprotein (VLDL), low-density lipoprotein (LDL) and high-density lipoprotein particle sizes) was assessed by high-performance liquid chromatography (HPLC) as described previously (Krupkova et al., 2010; Usui et al., 2002). Serum free fatty acids were determined using an acyl-CoA oxidase-based colorimetric kit (Roche Diagnostics GmbH, Mannheim, Germany). Serum insulin concentration was determined using an ELISA kit for rat insulin assay (Mercodia, Uppsala, Sweden). Serum levels of adiponectin were determined using Rat Adiponectin ELISA kit (B-Bridge International, Cupertino, CA, USA).
Transcriptomic profiling and quantitative real-time PCR
Total RNA was extracted with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and purified with the RNeasy MinElute cleanup kit (Qiagen, Valencia, CA, USA) following the manufacturer's recommendations. The quality of the total RNA was evaluated on an Agilent 2100 Bioanalyzer system (Agilent, Palo Alto, CA, USA). Microarray experiments were performed using the GeneChip Rat Exon 1.0 ST array (interrogates over 850 000 exon clusters), with approximately four probes per exon and roughly 40 probes per gene (Affymetrix, Santa Clara, CA, USA). Following the ribosomal reduction procedure (Invitrogen), from 1 μg total RNA, each sample was processed using the GeneChip Whole Transcript Sense Target Labeling Assay (Affymetrix). Briefly, after the rRNA reduction procedure, double-stranded cDNA was synthesized with random hexamers tagged with a T7 promoter sequence, then it was amplified by T7 RNA polymerase producing complementary RNA; in the second cycle of cDNA synthesis, complementary RNA was used to produce sense single-stranded cDNA and 5.5 μg was fragmented, labeled and hybridized onto the chip (PD.SHR4a–2 chips; PD–3 chips). The whole hybridization procedure was performed using the Affymetrix GeneChip system according to the protocol recommended by Affymetrix. The hybridization was evaluated with Affymetrix GeneChip Command Console Software (AGCC) and the quality of the chips with Affymetrix Expression Console. Partek Genomics Suite (Partek, St Louis, MO, USA) was used for data analysis. The data were normalized by using Robust Multichip Average (RMA) algorithm, which uses background adjustment, quantile normalization and summarization.
To validate microarray gene expression data, quantitative real-time PCR (SYBR-Green) was used. Total RNA (2 μg) was reverse-transcribed with random primers using the SuperScript VILO cDNA Synthesis Kit (Invitrogen). Primers were designed using Primer3 (Rozen and Skaletsky, 2000) and synthesized by Integrated DNA Technologies. The primer sequences are listed in Supplementary Table 2. Real-time PCR reaction was performed in quadruplicate with EXPRESS SYBR GreenER qPCR SuperMix with Premixed ROX Kit according to the manufacturer's protocol (Invitrogen, Burlington, ON, Canada) using Applied Biosystems (Burlington, ON, Canada) Real-Time PCR System. Results were analyzed using the Pfaffl analysis method (Pfaffl, 2001) with reference genes peptidylprolyl isomerase B and 18S.
Statistical and pathway analyses
The metabolic and morphometric data were compared by one-way analysis of variance with strain as main factor followed by Tukey's honest significance difference test for detailed pair-wise comparison.
Transcriptomic data
After evaluation of the hybridization, the quality control and the data normalization by robust multi-array analysis (RMA), the gene expression was compared between the PD.SHR4a and PD strains using Partek Genomics Suite (Partek). The transcripts found to be significantly differentially expressed between PD and PD.SHR4a strains (FDR<0.1, n=172) were included in the gene enrichment and pathway/network analyses, which were performed using web-based DAVID functional enrichment analysis (Huang da et al., 2009) and Ingenuity Pathway Analysis software v.9 applications.
Results
Genomic characteristics of the differential segments in the new PD.SHR4 congenic strains
Our genotyping scan of 54 polymorphic microsatellite markers revealed the extent of the chromosome 4 differential segments of SHR origin in the two PD.SHR4 congenic strains (Table 1). In PD.SHR(D4Arb13-D4Rat125)—PD.SHR4a hereafter—the SHR-derived segment spans about 21 Mb, while in PD.SHR(cen-Lmbr; D4Rat147–D4Rat151)—PD.SHR4b hereafter—the introgressed segment is discontinuous as it extends 37 Mb from the centromere with a limited interruption at 1.3–2.4 Mb (Table 1). Several total genome scans conducted throughout the PD.SHR4 strains derivation eventually excluded the presence of other non-PD alleles than those fixed on chromosome 4, confirming the congenicity of the new strains. The SHR-derived segments on chromosome 4 hence represent the only genomic differences between PD and PD.SHR4 congenic strains.
Morphometry and basic metabolic profile
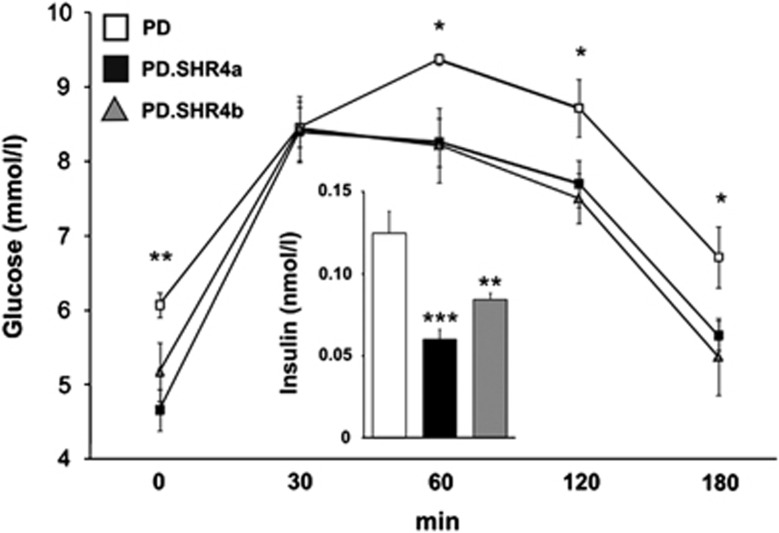
The PD and the two PD.SHR4 strains did not show any differences in body weight, adipose tissue distribution or relative weights of internal organs except for the slightly lower liver weight in PD.SHR4a (Table 2). Both congenic strains displayed significantly lower levels of fasting plasma glucose and insulin, as well as an overall amelioration of glucose tolerance compared with the PD progenitor (Figure 1). The concentrations of plasma adiponectin and free fatty acids were comparable among the three strains, while free glycerol was significantly lower in both PD.SHR4a and PD.SHR4b congenics (Table 2).
Table 2. Morphometric profile of PD versus PD.SHR4 congenic rats.
| Trait | PD | PD.SHR4a | PD.SHR4b | PANOVA |
|---|---|---|---|---|
| N | 8 | 8 | 6 | |
| Body weight, g | 317±6 | 313±11 | 297±9 | 0.18 |
| Liver wt., g per 100 g b.wt. | 3.11±0.04 | 2.93±0.02 | 3.13±0.02a | 0.004 |
| Heart wt., g per 100 g b.wt. | 0.29±0.002 | 0.29±0.01 | 0.28±0.01 | 0.96 |
| Kidney wt., g per 100 g b.wt. | 0.62±0.01 | 0.61±0.01 | 0.63±0.01 | 0.35 |
| Adrenals wt., mg per 100 g b.wt. | 17.9±0.8 | 16.7±0.2 | 17.3±0.5 | 0.17 |
| EFP wt., g per 100 g b.wt. | 1.45±0.12 | 1.36±0.12 | 1.36±0.07 | 0.79 |
| RFP wt., g per 100 g b.wt. | 1.37±0.17 | 1.29±0.13 | 1.08±0.16 | 0.54 |
| FFA, mmol l−1 | 0.75±0.01 | 0.80±0.04 | 0.72±0.05 | 0.29 |
| Fasting adiponectin, μg ml−1 | 3.2±0.6 | 3.5±0.6 | 4.0±1.0 | 0.77 |
| Free glycerol, mg dl−1 | 7.4±0.3 | 5.1±0.2‡ | 5.2±0.4† | 0.0004 |
Abbreviations: b.wt., body weight; EFP, epidymal fat pad; FFA, free faty acids; PD, polydactylous; RFP, retroperitoneal fat pad; SHR, spontaneously hypertensive rat.
Morphometric profile of PD versus PD.SHR4a and PD.SHR4b male rats fed standard diet. Values are shown as mean±s.e.m. The significance levels of pair-wise, inter-strain comparisons between PD.SHR4 congenic strains versus PD are shown for post-hoc Tukey's HSD test as follows: †P<0.01; ‡P<0.001 and aP<0.05 for the differences between PD.SHR4a and PD.SHR4b. Bold values indicate significant ANOVA result, italic values indicate non-significant ANOVA results.
Figure 1.
The glucose tolerance and fasting insulin in PD vs PD.SHR4 congenic strains. The course of glycemic curves in PD (white squares) vs PD.SHR4a (black squares) and PD.SHR4b (gray triangles) male rats fed standard diet. Fasting insulin concentrations in PD (white bar) vs PD.SHR4a (black bar) and PD.SHR4b (gray bar). Significance levels for oral glucose-tolerance test are given for the factor strain of one-way ANOVA; for fasting insulin, the strain comparison using the post-hoc Tukey's honest significance difference test of the one-way ANOVA with strain as major factor is shown as follows: *P<0.05; **P<0.01; ***P<0.001.
Detailed lipid profile
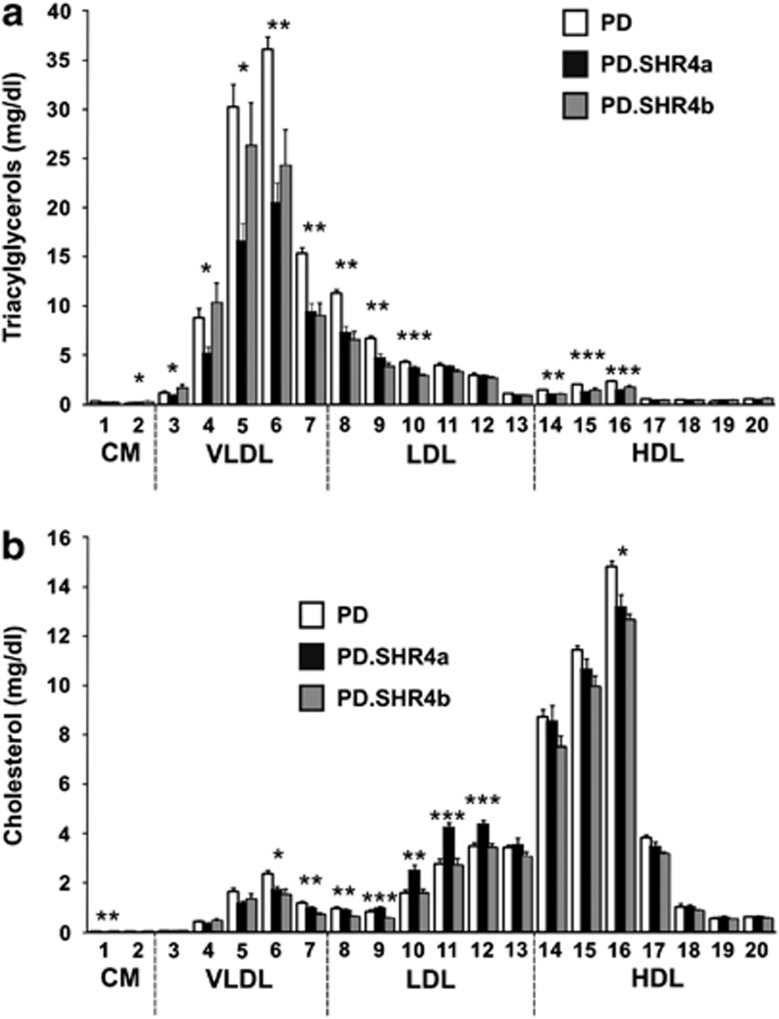
We observed significantly lower total triglyceride (TG) concentration in PD.SHR4a compared to PD, particularly driven by 43% and 22% differences in VLDL and LDL fractions, respectively (Table 3, Figure 2a). The same, yet less pronounced trend towards lower TG was present in PD.SHR4b, reaching statistical significance only for LDL and high-density lipoprotein fractions (Table 3, Figure 2a). While the total cholesterol was comparable between PD and PD.SHR4a congenic strain, upon a detailed analysis we observed significant elevation of LDL cholesterol in the congenic, unique to this strain (Table 3, Figure 2b). Total cholesterol was lower in PD.SHR4b compared to PD, mostly due to lower concentrations of high-density lipoprotein cholesterol (Table 3, Figure 2b). Only PD.SHR4b showed somewhat altered sizes of lipoprotein particles with larger VLDL compared to both PD and PD.SHR4a and smaller LDL particles compared to PD.SHR4a only (Supplementary Table 1).
Table 3. Triacylglycerol and cholesterol concentrations in major lipoprotein subfractions in PD vs PD.SHR4 congenic rats.
| Trait (mg dl−l) | PD | PD.SHR4a | PD.SHR4b | PANOVA |
|---|---|---|---|---|
| N | 8 | 8 | 6 | |
| Total TG | 130.3±5.7 | 82.2±7.2* | 98.6±12.7 | 0.017 |
| Chylomicron TG | 0.41±0.04 | 0.42±0.05 | 0.46±0.07 | 0.83 |
| VLDL-TG | 91.8±6.0 | 52.5±5.6* | 71.7±11.2 | 0.022 |
| LDL-TG | 30.4±1.4 | 23.6±1.6* | 20.2±1.0† | 0.001 |
| HDL-TG | 7.7±0.3 | 5.7±0.2† | 6.2±0.5* | 0.008 |
| Cholesterol (C) | ||||
| Total C | 60.0±1.5 | 59.0±1.8 | 51.4±1.6*,a | 0.009 |
| Chylomicron C | 0.03±0.003 | 0.03±0.01 | 0.06±0.01a | 0.019 |
| VLDL-C | 5.7±0.4 | 4.3±0.2 | 4.1±0.6 | 0.07 |
| LDL-C | 13.1±0.7 | 16.6±0.7*,b | 12.0±0.6 | 0.0006 |
| HDL-C | 41.1±0.8 | 38.2±1.5 | 35.3±1.1* | 0.035 |
Abbreviations: PD, polydactylous; SHR, spontaneously hypertensive rat.
Data are shown as mean±s.e.m. The significance levels of pair-wise, inter-strain comparisons between PD.SHR4 congenic strains and PD are shown for post-hoc Tukey's HSD test as follows: *P<0.05, †P<0.01, aP<0.05 and bP<0.001, for the differences between PD.SHR4a and PD.SHR4b. Bold values indicate significant ANOVA result, italic values indicate non-significant ANOVA results.
Figure 2.
(a) The triacylglycerols (a) and (b) cholesterol content in 20 lipoprotein subfractions in PD (white bars) versus PD.SHR4a (black bars) and PD.SHR4b (gray bars) male rats. Within the graph, the significance levels for strain differences are shown for one-way ANOVA (STRAIN factor) as follows: *P<0.05; **P<0.01; ***P<0.001. The allocation of individual lipoprotein subfractions to major lipoprotein classes is shown in order of the particle's decreasing size from left to right. CM, chylomicron; HDL, high-density lipoprotein; LDL, low-density lipoprotein; VLDL, very low-density lipoprotein.
Liver transcriptome comparison
As the PD.SHR4a strain carries a substantially smaller chromosome 4 differential segment compared to PD.SHR4b (Table 1) and at the same time it displayed more profound shifts in its metabolic profile, we decided to contrast its hepatic transcriptome with that of the PD rat. After filtering the results for quality parameters and adjusting statistical significance levels for multiple comparisons (see Materials and methods), we found 172 transcripts to be differentially expressed between PD and PD.SHR4a (Supplementary Data Set), with 48 of the transcripts showing more than a twofold difference (Table 4a). The numbers of transcripts up- and downregulated in PD.SHR4a vs PD were roughly balanced (83 up- vs 89 downregulated). Apart from the expected substantially reduced expression of Cd36 in the congenic strain, there was no other differentially expressed gene located in the segment of chromosome 4 of SHR origin (Table 4 and Supplemental Data Set). The rest of the differentially expressed genes were spread across almost all the other chromosomes. In order to validate the results of the microarray experiment, we performed quantitative real-time PCR assessment of the expression of nine representative genes: ATP-binding cassette, sub-family G (WHITE), member 5 (Abcg5); early growth response 1 (Egr1); insulin-like growth factor binding protein 2 (Igfbp2); period homolog 2 (Drosophila) (Per2); nuclear-receptor subfamily 1, group I, member 3 (Nr1i3); apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 (Apobec1); apolipoprotein L, 3 (Apol3); insulin induced gene 1 (Insig1); and aryl hydrocarbon receptor nuclear translocator-like (Arntl). As evident from Supplementary Figure 1, in all cases we were able to confirm the direction of expression change between PD and PD.SHR4a, in most of them the degree of up- or downregulation corresponded well. The relative overexpression of Igfbp2, Per2 and Nr1i3 in PD.SHR4a was even greater than the one ascertained by microarray.
Table 4. List of significantly differentially expressed transcripts between PD and PD.SHR4a (FDR<0.1, fold change >2). (a) Genes significantly overexpressed in PD.SHR4 vs PD (fold change >2); (b) genes significantly underexpressed in PD.SHR4 vs PD (fold change >2).
| Gene symbol | Gene name | Chr. | P-value | Fold change (PD vs PD.SHR4) |
|---|---|---|---|---|
| (a) | ||||
| Egr1 | Early growth response 1 | 18q | 1.28E−03 | 17.7 |
| Nat8 | N-acetyltransferase 8 (GCN5-related, putative) | 4q34 | 1.82E−03 | 11.5 |
| Abcg5 | ATP-binding cassette, sub-family G (WHITE), member 5 | 6q12 | 1.82E−04 | 11.2 |
| Igfbp2 | Insulin-like growth factor binding protein 2 | 9q33 | 1.04E−04 | 6.5 |
| Per3 | Period homolog 3 (Drosophila) | 5q36 | 1.01E−03 | 6.5 |
| Nr1i3 | nuclear receptor subfamily 1, group I, member 3 | 13q24 | 1.47E−03 | 5.7 |
| Por | P450 (cytochrome) oxidoreductase | 12q12 | 5.62E−04 | 4.8 |
| Per2 | Period homolog 2 (Drosophila) | 9q36 | 1.74E−03 | 4.6 |
| Aldh1a1 | Aldehyde dehydrogenase family 1, member A1 | 1q51 | 3.16E−03 | 4.6 |
| Gcnt2 | Glucosaminyl (N-acetyl) transferase 2, I-branching enzyme | 17p12 | 3.49E−04 | 4.0 |
| Cyp2j2 | Cytochrome P450, family 2, subfamily J, polypeptide 2 | 5q33 | 8.50E−04 | 3.6 |
| Pdgfc | Platelet-derived growth factor, C polypeptide | 2q33 | 3.78E−03 | 2.8 |
| Abcb1a | ATP-binding cassette, sub-family B (MDR/TAP), member 1 | 4q12 | 1.08E−03 | 2.6 |
| Ces2b/Ces2c | carboxylesterase 2C | 1q55 | 7.83E−04 | 2.6 |
| Tmem8 | Rattus norvegicus transmembrane protein 48 | 10q12 | 1.85E−04 | 2.5 |
| Itpr1 | Inositol 1,4,5-triphosphate receptor, type 1 | 4q41 | 1.62E−03 | 2.4 |
| Abo | ABO blood group (transferase A, alpha 1-3-N-acetylgalactosaminyltransferase; transferase B, alpha 1-3-galactosyltransferase) | 3p13 | 2.12E−03 | 2.4 |
| Abcd2 | ATP-binding cassette, sub-family D (ALD), member 2 | 7q35 | 9.16E−05 | 2.3 |
| C9orf95 | Chromosome 9 open reading frame 95 | 9 | 6.76E−04 | 2.3 |
| Herpud1 | Homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 | 19p12 | 1.39E−03 | 2.1 |
| Pnpla7 | Patatin-like phospholipase domain containing 7 | 3p13 | 2.00E−03 | 2.1 |
| (b) | ||||
| Arntl | Aryl hydrocarbon receptor nuclear translocator-like | 1q34 | 1.72E−03 | −13.5 |
| Cd36 | Cd36/Fatty acid translocase | 4q11 | 3.08E−04 | −11.7 |
| Insig1 | insulin induced gene 1 | 4q11 | 1.87E−03 | −10.0 |
| Phf11 | PHD finger protein 11 | 15p12 | 1.76E−03 | −9.6 |
| Acpp | Acid phosphatase, prostate | 8q32 | 5.32E−04 | −7.0 |
| Apol3 | Apolipoprotein L, 3 | 7q34 | 2.37E−04 | −6.9 |
| Srd5a1 | Steroid-5-alpha-reductase, alpha polypeptide 1 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 1) | 17p14 | 9.26E−04 | −6.7 |
| Slc34a2 | Solute carrier family 34 (sodium phosphate), member 2 | 14q11 | 7.86E−04 | −6.1 |
| Ltc4s | Leukotriene C4 synthase | 10q22 | 1.05E−03 | −5.7 |
| LOC360228 | WDNM1 homolog | 10q26 | 5.29E−04 | −5.3 |
| Nfil3 | Nuclear factor, interleukin 3 regulated | 17p14 | 3.00E−03 | −4.7 |
| Psmb8 | Proteasome (prosome, macropain) subunit, beta type 8 (large multifunctional peptidase 7) | 20p12 | 1.89E−03 | −3.1 |
| RGD1304580 | Similar to Hypothetical protein MGC38513 | 1q21 | 1.35E−03 | −2.9 |
| Cxadr | Coxsackie virus and adenovirus receptor | 11q11 | 4.83E−04 | −2.7 |
| Capg | capping protein (actin filament), gelsolin-like | 4q33 | 3.78E−03 | −2.7 |
| Oprs1 | Opioid receptor, sigma 1 | 5q22 | 1.98E−03 | −2.6 |
| Hamp | Hepcidin antimicrobial peptide | 1q21 | 2.43E−03 | −2.5 |
| Casp1 | Caspase 1 | 8q11 | 3.25E−03 | −2.5 |
| C6 | Complement component 6 | 2q16 | 1.01E−03 | −2.5 |
| Trim5 | Tripartite motif-containing 5 | 1q32 | 9.38E−04 | −2.3 |
| Tspo | Translocator protein | 7q34 | 3.56E−04 | −2.3 |
| Apobec1 | Apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 | 4q42 | 3.07E−03 | −2.3 |
| Bst2 | Bone marrow stromal cell antigen 2 | 16p14 | 8.53E−04 | −2.2 |
| Pgrmc2 | Progesterone receptor membrane component 2 | 2q26 | 3.58E−03 | −2.2 |
| Vwa5a | Von Willebrand factor A domain containing 5A | 8q22 | 3.16E−03 | −2.1 |
| Gtf2ird1 | GTF2I repeat domain containing 1 | 12q12 | 4.54E−05 | −2.0 |
| Anxa3 | Annexin A3 | 14p22 | 1.32E−05 | −2.0 |
The P-value is adjusted for multiple comparisons.
Pathway/network analysis
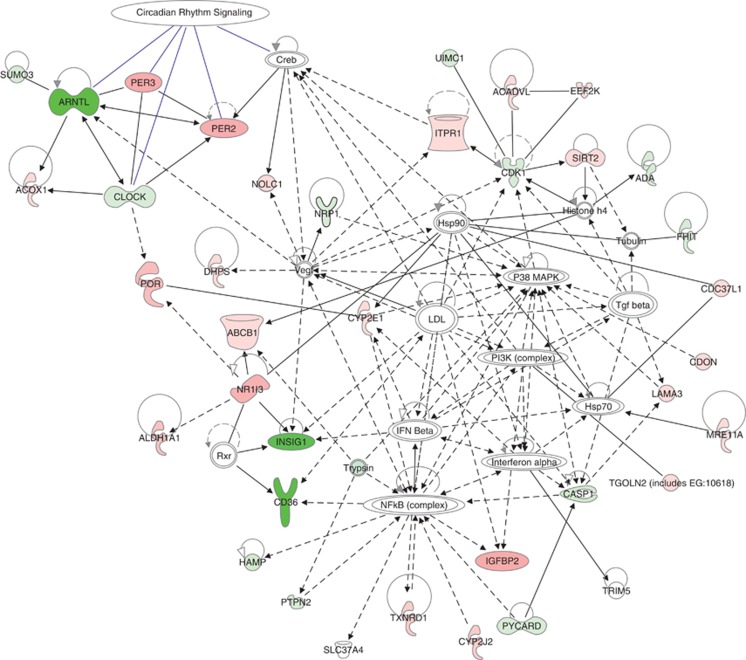
Using all 172 significantly differentially expressed genes, we carried out a systematic search for their enrichment in ontological categories, canonical pathways or disease-related gene sets as well as their potential functional connections. First, we examined the degree of over-representation of our set of genes in the canonical pathways using the relevant module in the Ingenuity Pathway Analysis v.9 software. After correction for multiple testing using Benjamini–Hochberg false discovery rate, we identified four pathways significantly enriched by the genes most distinctly expressed in PD vs PD.SHR4: circadian rhythm signaling, xenobiotic metabolism signaling, PXR/RXR activation and LPS/IL-1-mediated inhibition of RXR function (Supplementary Figure 2). In the subsequent toxicity functions analysis (as implemented in IPA), only liver regeneration, liver steatosis and renal tubule injury surpassed the statistical threshold for significant over-representation both in the total sample of 172 genes and in the subset upregulated in PD.SHR4. Then, we proceeded to assess the potential functional relations among the genes differentially expressed between PD.SHR4a and PD using dynamic pathway modeling. Both approaches focusing on direct interactions of identified genes or the ‘shortest path' among them revealed several major modules, the most prominent being formed by the four major genes involved in circadian rhythmicity (Figure 3).
Figure 3.
Network analysis of the differentially expressed genes in the livers of PD and PD.SHR4a strains. The figure represents network with the highest score (IPA v.9, Ingenuity Systems) derived using the whole set of differentially expressed genes between PD and PD.SHR4a. The genes relatively downregulated in PD.SHR4a vs PD are shown in shades of green; the genes relatively upregulated in PD.SHR4a vs PD are shown in shades of red (in both cases, the darker the color, the bigger the fold change of expression). The members of the circadian rhythm signaling group are indicated by blue arrows.
Discussion
The transfer of a limited segment of rat chromosome 4 including the mutated Cd36 gene of SHR origin into the genomic background of PD rat strain elicited an improvement in glucose tolerance, lowering of insulinaemia and shifts in lipid levels contrasting with those reported previously after a similar transfer to Brown Norway genomic background (Seda et al., 2002, 2003a; Table 5). As we have previously shown the concentrations of insulin, glucose, free fatty acids and cholesterol to be comparable in PD and SHR strains (Sedova et al., 2000), the current results indicate the presence of an interaction between gene(s) within the introgressed segments and genomic background of the congenic strains.
Table 5. Comparison of effect of Cd36 deficiency in different genetic models.
| BN.SHR4 vs BN | PD.SHR4 vs PD | Cd36−/− mice vs WT | |
|---|---|---|---|
| Fasting glucose | ↑ | ↓ | ↓ |
| Fasting insulin | ↑ | ↓ | ↓ |
| Fasting TG | ↑ | ↓ ↔ | ↑ |
| Fasting FFA | ↑ | ↔ | ↑ |
Abbreviations: FFA, free fatty acids;TG, triacylglycerols;
Summary of metabolic effects owing to introduction of mutant Cd36 into different genetic backgrounds. All data are based on reports utilizing standard chow only. The arrows indicate the respective change of the metabolic parameter in the Cd36-deficient animals versus their wild-type controls: ↑, increase; ↓, decrease; ↔, no change.
Although many of the studies including ours demonstrated the association of lack of Cd36 with metabolic syndrome features like insulin resistance or dyslipidemia (Aitman et al., 1997, 1999; Febbraio et al., 1999; Pravenec et al., 2001; Seda et al., 2002), there are also reports suggesting that, in certain circumstances, the expression level of Cd36 actually correlates positively with unfavorable metabolic profile. Recently, it has been shown by Love-Gregory et al. (2011) that in African-Americans the Cd36 variants that reduce protein expression appear to promote a protective metabolic profile, particularly concerning high-density lipoprotein and VLDL lipid levels (Love-Gregory et al., 2011). Also, the Cd36 knockout mice fed chow diet displayed lower fasting concentrations of glucose and insulin compared with wild-type controls (Hajri et al., 2002), similarly to PD.SHR4 congenics in the current study. On the other hand, their triglyceride and free fatty acid levels were increased in contrast to the decrease or no change observed in our study (Table 5). Absence of Cd36 also protects mice from insulin resistance associated with high fat diet-induced obesity and hyperlipidemia (Kennedy et al., 2011) and, conversely, increased expression of hepatic Cd36 in response to high fat diet-induced obesity was found to be sufficient to exacerbate hepatic triglyceride storage and secretion (Koonen et al., 2007). One of the mechanisms responsible may lie in the fact that human islets express Cd36 in the plasma membrane as well as in the insulin-secretory granules, and Cd36 activity was deemed important for uptake of fatty acids into β-cells as well as for mediating their modulatory effects on insulin secretion (Noushmehr et al., 2005) .
Altogether, it is apparent that the eventual metabolic effect of Cd36 deficiency is tightly linked to a particular setting of both genomic background (for example, PD.SHR4 vs BN.SHR4 (Seda et al., 2002, 2003b); Table 5 and Supplementary Figure 3) and environmental factors, particularly diet (Febbraio et al., 1999; Hajri et al., 2002; Koonen et al., 2007; Kennedy et al., 2011) or medication (Qi et al., 2002; Seda et al., 2003a; Seda et al., 2008; Krupkova et al., 2010). Therefore, the apparently controversial issue of causal relation between level of Cd36 expression and metabolic outcome may be resolved by adoption of broader conceptual framework incorporating other (eco)genomic factors.
Apart from Cd36, significant overexpression of Igfbp2 in PD.SHR4a (yet genetically of PD origin in both strains) represents a possible mediator of the enhanced insulin sensitivity, as was reported, for example, in Igfbp2 transgenic mice (Wheatcroft et al., 2007). The observation of concurrent upregulation of Abcg5 and downregulation of Insig1 in PD.SHR4 congenic follows well their assumed respective roles in cholesterol efflux and its cellular feedback regulation. This shift, reported also in Insig1−/−Insig2−/− mice (Engelking et al., 2005), may suggest a tendency toward increasing lipid deposition in liver, although we did not validate this parameter directly.
One of the intriguing results in the current study was the identification of a differentially expressed set of circadian clock-related genes (Figure 3). The fundamental interrelatedness of circadian rhythmicity, inter- and intra-organ desynchrony with the metabolic and signaling pathways involved in the pathogenesis of cardiometabolic diseases has been supported by an ever-growing body of evidence, reviewed recently in detail elsewhere (Eckel-Mahan-Sassone-Corsi, 2009; Maury et al., 2010). The gene most downregulated in PD.SHR4a liver compared with PD was Arntl (also known as Bmal1), an essential component of master circadian pacemaker (Bunger et al., 2000), together with upregulated Per2 and Per3 genes, two natural inhibitors of Bmal1 and parts of its negative-feedback loop. There is a certain resemblance to the studies in liver-specific Bmal1 knockout mice, which show hypoglycemia restricted to the fasting phase of the daily cycle and enhanced glucose clearance after an overnight fast (Lamia et al., 2008), but not reduced fasting-insulin levels, found in both PD.SHR4 congenic strains and complete Bmal1 knockout (Lamia et al., 2008). We are not aware of any previous report functionally linking the metabolic outcomes of Cd36 polymorphisms to circadian-clock circuitry.
Finally, we observed several differences in lipid levels and liver weight between the two congenic strains. There are several plausible candidates for these effects within the three regions differing between PD.SHR4a and PD.SHRb, namely Insig1, serotonin receptor Htr5a or carnitine O-octanoyltransferase (Crot) as indicated in Table 1. We cannot completely exclude the possibility that some of the other genes of SHR origin are present in the differential segment(s) of the congenic PD.SHR4 strains contributed to the observed metabolic effects commonly present in both congenics and SHR. However, the transcriptome analysis and functional annotation of all the genes in the segment reinforce rather the effect of the Cd36 mutation. Within the differential segment of the PD.SHR4a strain, there are 58 well-documented protein-coding genes (Supplemental Table 3) among about 120 transcripts. Of those, only Cd36 was found to be differentially expressed between PD and PD.SHR4a. Further studies are needed to assess the effect of Cd36 and the whole differential segment under distinct nutritional and pharmacological challenges, especially given our previous extensive documentation of nutrigenetic and pharmacogenetic interactions of a similar genomic region in the BN.SHR4 congenic strain (Seda et al., 2003a, 2008; Krupkova et al., 2010). In sum, the introduction of mutated Cd36 into the genomic background of an inbred model of metabolic syndrome resulted in disconnected shifts of metabolic profile along with distinct changes in hepatic transcriptome. Our network analysis revealed possible pathways underlie the improvement of insulin sensitivity toward a shift in lipid profile. The synthesis of the current results with those obtained in other Cd36-deficient strains indicates that the eventual metabolic effect of a deleterious mutation such as that of SHR-derived Cd36 is not absolute, but rather a function of complex interactions between environment and genomic background, upon which it operates.
Data archiving
Complete microarray datasets have been submitted to the NCBI Gene Expression Omnibus (GEO) under the record GSE33597. Remaining data have been deposited in the Dryad repository; doi:10.5061/dryad.9f8q91q3.
Acknowledgments
We thank Michaela Janků, Barbora Baladová and Marie Uxová for their excellent technical assistance. This work was supported by Research Project MSM 0021620807 and by the scientific programme of Charles University, PRVOUK-P25/LF1/2.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Hypertension Research website (http://www.nature.com/hr)
Supplementary Material
References
- Aitman TJ, Glazier AM, Wallace CA, Cooper LD, Norsworthy PJ, Wahid FN, et al. Identification of Cd36 (Fat) as an insulin-resistance gene causing defective fatty acid and glucose metabolism in hypertensive rats. Nat Genet. 1999;21:76–83. doi: 10.1038/5013. [DOI] [PubMed] [Google Scholar]
- Aitman TJ, Gotoda T, Evans AL, Imrie H, Heath KE, Trembling PM, et al. Quantitative trait loci for cellular defects in glucose and fatty acid metabolism in hypertensive rats. Nat Genet. 1997;16:197–201. doi: 10.1038/ng0697-197. [DOI] [PubMed] [Google Scholar]
- Alberti KGMM, Eckel RH, Grundy SM, Zimmet PZ, Cleeman JI, Donato KA, et al. Harmonizing the Metabolic Syndrome: A Joint Interim Statement of the International Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation. 2009;120:1640–1645. doi: 10.1161/CIRCULATIONAHA.109.192644. [DOI] [PubMed] [Google Scholar]
- Bokor S, Legry V, Meirhaeghe A, Ruiz JR, Mauro B, Widhalm K, et al. Single-nucleotide polymorphism of CD36 locus and obesity in European adolescents. Obesity (Silver Spring) 2010;18:1398–1403. doi: 10.1038/oby.2009.412. [DOI] [PubMed] [Google Scholar]
- Bonen A, Han XX, Tandon NN, Glatz JF, Lally J, Snook LA, et al. FAT/CD36 expression is not ablated in spontaneously hypertensive rats. J Lipid Res. 2009;50:740–748. doi: 10.1194/jlr.M800237-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, et al. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choquet H, Labrune Y, De Graeve F, Hinney A, Hebebrand J, Scherag A, et al. Lack of association of CD36 SNPs with early onset obesity: a meta-analysis in 9,973 European subjects. Obesity (Silver Spring) 2010;19:833–839. doi: 10.1038/oby.2010.226. [DOI] [PubMed] [Google Scholar]
- Dwinell MR, Worthey EA, Shimoyama M, Bakir-Gungor B, DePons J, Laulederkind S, et al. The Rat Genome Database 2009: variation, ontologies and pathways. Nucleic Acids Res. 2009;37:D744–749. doi: 10.1093/nar/gkn842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel-Mahan K, Sassone-Corsi P. Metabolism control by the circadian clock and vice versa. Nat Struct Mol Biol. 2009;16:462–467. doi: 10.1038/nsmb.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelking LJ, Liang G, Hammer RE, Takaishi K, Kuriyama H, Evers BM, et al. Schoenheimer effect explained--feedback regulation of cholesterol synthesis in mice mediated by Insig proteins. J Clin Invest. 2005;115:2489–2498. doi: 10.1172/JCI25614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Febbraio M, Abumrad NA, Hajjar DP, Sharma K, Cheng W, Pearce SF, et al. A null mutation in murine CD36 reveals an important role in fatty acid and lipoprotein metabolism. J Biol Chem. 1999;274:19055–19062. doi: 10.1074/jbc.274.27.19055. [DOI] [PubMed] [Google Scholar]
- Glazier AM, Scott J, Aitman TJ. Molecular basis of the Cd36 chromosomal deletion underlying SHR defects in insulin action and fatty acid metabolism. Mamm Genome. 2002;13:108–113. doi: 10.1007/s00335-001-2132-9. [DOI] [PubMed] [Google Scholar]
- Hajri T, Han XX, Bonen A, Abumrad NA. Defective fatty acid uptake modulates insulin responsiveness and metabolic responses to diet in CD36-null mice. J Clin Invest. 2002;109:1381–1389. doi: 10.1172/JCI14596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Kashiwagi H, Tomiyama Y, Nozaki S, Honda S, Kosugi S, Shiraga M, et al. A Single Nucleotide Insertion in Codon 317 of the CD36 Gene Leads to CD36 Deficiency. Arterioscler Thromb Vasc Biol. 1996;16:1026–1032. doi: 10.1161/01.atv.16.8.1026. [DOI] [PubMed] [Google Scholar]
- Kennedy DJ, Kuchibhotla S, Westfall KM, Silverstein RL, Morton RE, Febbraio M. A CD36-dependent pathway enhances macrophage and adipose tissue inflammation and impairs insulin signalling. Cardiovasc Res. 2011;89:604–613. doi: 10.1093/cvr/cvq360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonen DPY, Jacobs RL, Febbraio M, Young ME, Soltys C-LM, Ong H, et al. Increased hepatic CD36 expression contributes to dyslipidemia associated with diet-induced obesity. Diabetes. 2007;56:2863–2871. doi: 10.2337/db07-0907. [DOI] [PubMed] [Google Scholar]
- Kraja AT, Vaidya D, Pankow JS, Goodarzi MO, Assimes TL, Kullo IJ, et al. A bivariate genome-wide approach to metabolic syndrome: STAMPEED Consortium. Diabetes. 2011;60:1329–1339. doi: 10.2337/db10-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kren V. Genetics of the polydactyly-luxate syndrome in the Norway rat, Rattus norvegicus. Acta Univ Carol Med Monogr. 1975. pp. 1–103. [PubMed]
- Krupkova M, Sedova L, Liska F, Krenova D, Kren V, Seda O. Pharmacogenetic interaction between dexamethasone and Cd36-deficient segment of spontaneously hypertensive rat chromosome 4 affects triacylglycerol and cholesterol distribution into lipoprotein fractions. Lipids Health Dis. 2010;9:38. doi: 10.1186/1476-511X-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci U S A. 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love-Gregory L, Sherva R, Schappe T, Qi JS, McCrea J, Klein S, et al. Common CD36 SNPs reduce protein expression and may contribute to a protective atherogenic profile. Hum Mol Genet. 2011;20:193–201. doi: 10.1093/hmg/ddq449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love-Gregory L, Sherva R, Sun L, Wasson J, Schappe T, Doria A, et al. Variants in the CD36 gene associate with the metabolic syndrome and high-density lipoprotein cholesterol. Hum Mol Genet. 2008;17:1695–1704. doi: 10.1093/hmg/ddn060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maury E, Ramsey KM, Bass J. Circadian rhythms and metabolic syndrome: from experimental genetics to human disease. Circ Res. 2010;106:447–462. doi: 10.1161/CIRCRESAHA.109.208355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel SE, Lai CQ, Mattei J, Parnell LD, Ordovas JM, Tucker KL. Variants of the CD36 gene and metabolic syndrome in Boston Puerto Rican adults. Atherosclerosis. 2010;211:210–215. doi: 10.1016/j.atherosclerosis.2010.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noushmehr H, D'Amico E, Farilla L, Hui H, Wawrowsky KA, Mlynarski W, et al. Fatty acid translocase (FAT/CD36) is localized on insulin-containing granules in human pancreatic beta-cells and mediates fatty acid effects on insulin secretion. Diabetes. 2005;54:472–481. doi: 10.2337/diabetes.54.2.472. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pravenec M, Landa V, Zidek V, Musilova A, Kren V, Kazdova L, et al. Transgenic rescue of defective Cd36 ameliorates insulin resistance in spontaneously hypertensive rats. Nat Genet. 2001;27:156–158. doi: 10.1038/84777. [DOI] [PubMed] [Google Scholar]
- Qi N, Kazdova L, Zidek V, Landa V, Kren V, Pershadsingh HA, et al. Pharmacogenetic evidence that cd36 is a key determinant of the metabolic effects of pioglitazone. J Biol Chem. 2002;277:48501–48507. doi: 10.1074/jbc.M206655200. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Seda O, Kazdova L, Krenova D, Kren V. Rosiglitazone fails to improve hypertriglyceridemia and glucose tolerance in CD36-deficient BN.SHR4 congenic rat strain. Physiol Genomics. 2003a;12:73–78. doi: 10.1152/physiolgenomics.00113.2002. [DOI] [PubMed] [Google Scholar]
- Seda O, Liska F, Krenova D, Kazdova L, Sedova L, Zima T, et al. Dynamic genetic architecture of metabolic syndrome attributes in the rat. Physiol Genomics. 2005;21:243–252. doi: 10.1152/physiolgenomics.00230.2004. [DOI] [PubMed] [Google Scholar]
- Seda O, Liska F, Krenova D, Kazdova L, Sedova L, Zima T, et al. Differential linkage of triglyceride and glucose levels on rat chromosome 4 in two segregating rat populations. Folia Biol (Praha) 2003b;49:223–226. [PubMed] [Google Scholar]
- Seda O, Sedova L, Kazdova L, Krenova D, Kren V. Metabolic characterization of insulin resistance syndrome feature loci in three brown Norway-derived congenic strains. Folia Biol (Praha) 2002;48:81–88. [PubMed] [Google Scholar]
- Seda O, Sedova L, Oliyarnyk O, Kazdova L, Krenova D, Corbeil G, et al. Pharmacogenomics of metabolic effects of rosiglitazone. Pharmacogenomics. 2008;9:141–155. doi: 10.2217/14622416.9.2.141. [DOI] [PubMed] [Google Scholar]
- Sedova L, Kazdova L, Seda O, Krenova D, Kren V. Rat inbred PD/cub strain as a model of dyslipidemia and insulin resistance. Folia Biol (Praha) 2000;46:99–106. [PubMed] [Google Scholar]
- Usui S, Hara Y, Hosaki S, Okazaki M. A new on-line dual enzymatic method for simultaneous quantification of cholesterol and triglycerides in lipoproteins by HPLC. J Lipid Res. 2002;43:805–814. [PubMed] [Google Scholar]
- Wheatcroft SB, Kearney MT, Shah AM, Ezzat VA, Miell JR, Modo M, et al. IGF-binding protein-2 protects against the development of obesity and insulin resistance. Diabetes. 2007;56:285–294. doi: 10.2337/db06-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto N, Akamatsu N, Sakuraba H, Yamazaki H, Tanoue K. Platelet glycoprotein IV (CD36) deficiency is associated with the absence (type I) or the presence (type II) of glycoprotein IV on monocytes. Blood. 1994;83:392–397. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.