Figure 4.
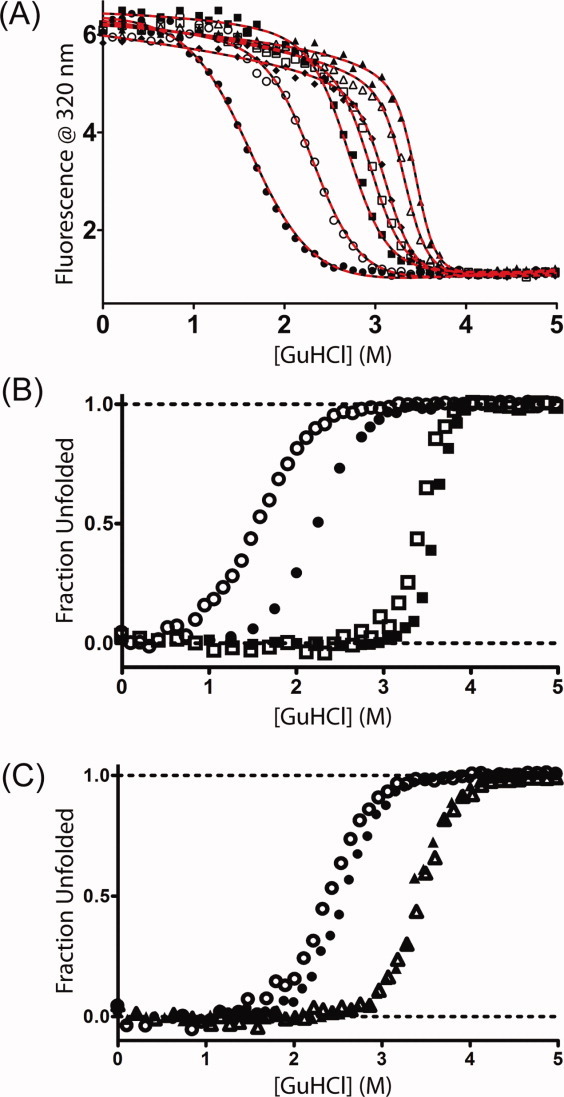
Denaturation of consensus TPR proteins in GuHCl at 25°C. (A) Fluorescence at 320 nm for CTPRan constructs with protein concentrations between 1 and 4 μM in 50 mM phosphate at pH 7.0—CTPRa2 (filled circles), CTPRa3 (open circles), CTPRa4 (filled squares), CTPRa5 (open squares), CTPRa6 (filled diamonds), CTPRa8 (open triangles), CTPRa10 (filled triangles). Lines correspond to the global best fits to a description based on the Regan-derived (solid black) or Barrick-derived (dashed red) one-dimensional Ising model—Materials and Methods. (B) Comparison of fraction unfolded versus [GuHCl] for CTPRa2 (circles) and CTPRa10 (squares) under differing buffer conditions: open symbols—50 mM phosphate, pH 7.0; filled symbols—50 mM phosphate, pH 6.5 + 150 mM NaCl (filled symbols—data obtained from Kajander et al.12 with the program Un-SCAN-IT [Silk Scientific]). (C) Comparison of fraction unfolded versus [GuHCl] for CTPR2 (circles) and CTPR3 (triangles) under differing buffer conditions: open symbols—50 mM phosphate, pH 7.0; filled symbols—pH 6.8 phosphate buffer + 150 mM NaCl (filled symbols—data were obtained from Main et al.23
