Background: Antisense mitochondrial ncRNAs are down-regulated during oncogenesis by unknown mechanisms.
Results: High risk HPV E2 oncogene induces down-regulation of the antisense transcripts. Additionally, E6 and E7 induce expression of a new sense mitochondrial ncRNA.
Conclusion: HPV oncogenes modulate expression of mitochondrial ncRNAs.
Significance: During non-viral oncogenesis, cellular factor(s), analogously to E2, could induce down-regulation of the antisense mitochondrial ncRNAs.
Keywords: Cancer, Mitochondria, Oncogene, Oncogenic Viruses, Papillomavirus, Non-coding RNA (ncRNA)
Abstract
The study of RNA and DNA oncogenic viruses has proved invaluable in the discovery of key cellular pathways that are rendered dysfunctional during cancer progression. An example is high risk human papillomavirus (HPV), the etiological agent of cervical cancer. The role of HPV oncogenes in cellular immortalization and transformation has been extensively investigated. We reported the differential expression of a family of human mitochondrial non-coding RNAs (ncRNAs) between normal and cancer cells. Normal cells express a sense mitochondrial ncRNA (SncmtRNA) that seems to be required for cell proliferation and two antisense transcripts (ASncmtRNAs). In contrast, the ASncmtRNAs are down-regulated in cancer cells. To shed some light on the mechanisms that trigger down-regulation of the ASncmtRNAs, we studied human keratinocytes (HFK) immortalized with HPV. Here we show that immortalization of HFK with HPV-16 or 18 causes down-regulation of the ASncmtRNAs and induces the expression of a new sense transcript named SncmtRNA-2. Transduction of HFK with both E6 and E7 is sufficient to induce expression of SncmtRNA-2. Moreover, E2 oncogene is involved in down-regulation of the ASncmtRNAs. Knockdown of E2 in immortalized cells reestablishes in a reversible manner the expression of the ASncmtRNAs, suggesting that endogenous cellular factors(s) could play functions analogous to E2 during non-HPV-induced oncogenesis.
Introduction
Cancer is characterized by a dysregulation of cell cycle control mechanisms, resulting in uncontrolled cell growth. Oncogenes and tumor suppressors, when functioning together properly, regulate progression of normal cell proliferation. In cancer, however, mutations result in constitutive activation of oncogenes, inactivation of tumor suppressors, immortality, resistance to apoptosis, invasiveness, and metastasis (1). About 15–20% of cancers are associated with infection by DNA and RNA oncogenic viruses (2–4), and the study of these pathogens has been invaluable in the discovery of key cellular pathways that become dysfunctional during cancer progression. For example, oncoproteins E6 and E7 from high risk human papillomavirus (HPV)3-16 or -18 disable tumor suppressors p53 and Rb and up-regulate telomerase, fundamental changes for cell immortalization (5, 6). Interestingly, an apparently important step in the induction of cancer by oncogenic viruses is the specific interaction of some viral oncogenes with mitochondria, an organelle that has been implicated for decades in carcinogenesis (7, 8).
Human cells express a unique family of sense and antisense mitochondrial ncRNAs containing long inverted repeats (IRs) (9, 10). The sense transcript or SncmtRNA (hereafter referred to as SncmtRNA-1), which contains an 815-nt IR and consequently a stem-loop structure (9), is expressed in normal proliferating cells and tumor cells but not in resting cells. This correlation between cell proliferation and expression of the SncmtRNA-1 suggests a function for this transcript in cell cycle progression (9, 10). Indeed, we have shown that rhodamine 6G, a drug that disables mitochondrial function (11, 12), inhibits in a reversible manner the expression of the SncmtRNA-1, together with DNA synthesis and PCNA expression in phytohemagglutinin-stimulated human lymphocytes (10). If the drug is removed, DNA synthesis and expression of the SncmtRNA-1 are reestablished. To fulfill this role, the transcript should be localized in the cytoplasm and/or nucleus. Recently, we showed that this mitochondrial transcript exits the organelle and localizes to the cytoplasm and the nucleus, where it can be found associated with chromatin and nucleoli (13).
In addition, normal proliferating cells express two antisense transcripts named ASncmtRNA-1 and -2 that contain IRs of 316 and 545 nt, respectively (10). In contrast, however, tumor cell lines as well as tumor cells present in human biopsies of different cancer types down-regulate the expression of the ASncmtRNAs (10). These observations suggest that down-regulation of the ASncmtRNAs is an essential step during the process of neoplastic transformation and progression. Therefore, one pending question is which cellular factor(s) is involved in down-regulation of the ASncmtRNAs during oncogenic transformation. This is indeed a difficult task, and as a first approach to addressing this question, we studied normal human foreskin keratinocytes (HFK) immortalized with high risk HPV-16 and -18 (14, 15). Here we show that the E2 oncogene from these viruses induces down-regulation of the ASncmtRNAs. On the other hand, E6 and E7 oncogenes induce the expression of a new sense transcript or SncmtRNA-2, which might be a potential biomarker to detect early intraepithelial neoplasia. In addition, we show that knockdown of SncmtRNA-1 induces inhibition of proliferation in SiHa and HeLa cells, suggesting a role for this transcript in cell cycle regulation.
EXPERIMENTAL PROCEDURES
Cell Culture
HeLa and SiHa cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) containing 10% fetal calf serum (FCS), 2 mm glutamine, 50 units/ml penicillin, 50 μg/ml streptomycin, and 0.1 mm non-essential amino acids (DMEM10). Pooled neonatal HFK (Lonza, CH) were cultured in keratinocyte serum-free medium (Invitrogen). HPV-16-immortalized cells (HFK698) and HPV-18-immortalized cells (18Nco and HF18) were cultured in keratinocyte serum-free medium/DMEM10 (3:1) (14, 15). All human cell lines were maintained in a humidified cell culture chamber at 37 °C and 5% CO2.
Retroviral Vectors and Primary HFK Cultures
Recombinant pLXSN retroviral vectors, either empty or containing HPV-16 E6, E7, or E6/E7 wild-type sequences were established as described elsewhere (16, 17). Low passage HFK were grown in serum-free medium (Invitrogen). Cells were acutely transduced with recombinant pLXSN vectors expressing the neomycin resistance marker. After 24 h, the cells were selected with 300 μg/ml G418 for 2 days, to eliminate non-transduced cells. Surviving cells were amplified and used to seed monolayer cultures.
RT-PCR Amplification
Total RNA from cells was extracted with TRIzol (Invitrogen) as described before (9, 10). To eliminate mtDNA contamination, RNA preparations were treated with TURBO DNA-free (Ambion) according to the manufacturer's instructions. Reverse transcription was carried out with 50–100 ng of freshly prepared RNA, 50 ng of random hexamers or sequence-specific primers, and 200 units of reverse transcriptase (Moloney murine leukemia virus, Invitrogen). The cDNA was PCR-amplified and analyzed by electrophoresis as described before (9, 10). Amplified DNA fragments were purified (Wizard SU Gel and PCR Clean-up system, Promega) and cloned in pGEM-T Easy (Promega) or pTOPO (Invitrogen), and the purified recombinant plasmids were sequenced as described before (9, 10). The primers used to amplify the SncmtRNA were deduced from the sequence of the human mitochondrial 16S rRNA (GenBankTM accession number V00662): P1 (r), 5′-AAGGTGGAGTGGGTTTGGGGC (positions 11–31); P2 (f), 5′-GGGGTCTTAGCTTTGGCTCTCC (positions 1326–1347); P3 (f), 5′-ACAACCAGCTATCACCAG (positions 333–353); P4 (f), 5′-TTGGTGGCTGCTTTTAGGCCTA (positions 1207–1227); P5 (f), 5′-TAGGGTGATAGATTGGTCCAA (positions 596–616); P6 (f), 5′-GGTTGATTGTAGATATTGGGCT (positions 833–854); P7 (f), 5′-GGTAAGATTTGCCGAGTTC (positions 741–59); P8 (r), 5′-AGTGATTAGCTACCTTTGCACGGT (positions 912–934); P9 (f), 5′-ACCGTGCAAAGGTAGCATAATCAC (positions 912–934); P10 (r), 5′-AATAGGATTGCGCTGTTATCCCTA (positions 1260–1283); P11 (r), 5′-ACCATTACCCAAATAAAGTATAG (positions 53–77), where (r) and (f) represent reverse and forward primers, respectively. The primers targeted to the linker region between the IR and the sense 16S mtRNA of the SncmtRNA-1 and SncmtRNA-2 were P12 (r) (5′-AGGTTTAGCCAAACCATT) and P13 (r) (5′-AGGTTTAGCACCGCAAGGG), respectively. Control 9-nt primers directed to the IRs of SncmtRNA-1 and SncmtRNA-2 were P14 (r) (5′-CAAACCATT) and P15 (r) (5′-ACCGCAAGG), respectively.
The sequences of the primers to amplify the gene products of HPV-16 or HPV-18 were deduced from the complete sequences of these viruses (GenBankTM accession numbers NC_001526 and NC_001357, respectively). The primer used for amplification were as follows: for HPV-16 E1 mRNA, HPV-16 E1 (f) (5′-GCATTGGACTTACACCCA-G) and HPV-16 E1 (r) (5′-TACGCAATTTTGGAGGCTCT); for HPV-16 E1^E4, HPV-16 E1^E4 (f) (5′-GATCCTGCAGGCAACGAAGT) and HPV-16 E1^E4 (r) (5′-TCCGGTGTCTGGCTCTGATC); for HPV-16 E2, HPV-16 E2 (f) (5′-AAGGCCAGAGAAATGGGATT) and HPV-16 E2 (r) (5′-CCATCAAACTGCACTTCCAC); for HPV-16 E5, HPV-16 E5 (f) (5′-CACAACATTACTGGCGTGCT) and HPV-16 E5 (r) (5′-AAAAAGCGTGCATGTGTATG); for HPV-16 E6, HPV-16 E6 (f) (5′-ATGCACAGAGCTGCAAACAA) and HPV-16 E6 (r) (5′-TTTGCTTTTCTTCAGGACACA); for HPV-16 E7, HPV-16 E7 (f) (5′-GAAACCCAGCTGTAATCATGC) and HPV-16 E7 (r) (5′-CTGAGAACAGATGGGGCACA); for HPV-18 E1, HPV-18 E1 (f) (5′-GCAGCACAGAAAACAGTGGA) and HPV-18 E1 (r) (5′-CGTACTGCCGCCACTACATA); for HPV-18 E1^E4, HPV-18 E1^E4 (f) (5′-ATGGCTGATCCAGAAGTACCAG) and HPV-18 E1^E4 (r) (5′-ACCGAGAAGTGGGTTGACAG); for HPV-18 E2, HPV-18 E2 (f) (5′-TCTTTGCAGCAAGGGAACAT) and HPV-18 E2 (r) (5′-CACAGGTAGCGGTTTTGTCC); for HPV-18 E4, HPV-18 E4 (f) (5′-ACGACACGGTATCCGCTACT) and HPV-18 E4 (r) (5′-ACCGAGAAGTGGGTTGACAG); for HPV-18 E5, HPV-18 E5 (f) (5′-TTGTGTATGCATGTATGTGTGC) and HPV-18 E5 (r) (5′-GCAGGGGACGTTATTACCAC); for HPV-18 E6, HPV-18 E6 (f) (5′-GATCTGTGCACGGAACTGAA) and HPV-18 E6 (r) (5′-TTTTTCTGCTGGATTCAACG); for HPV-18 E7, HPV-18 E7 (f) (5′-ATGCATGGACCTAAGGCAAC) and HPV-18 E6 (r) (5′-GATGCACACCACGGACAC).
RNase Digestion
About 1 μg of total RNA from HFK698 cells in 50 μl of 2× SSC was incubated with RNase A at a final concentration of 50 μg/ml for 15 s at room temperature (9, 10, 18). The digested RNA was extracted with phenol/chloroform, ethanol-precipitated, and centrifuged at 14,000 × g for 20 min at 4 °C. The RNA pellet was then washed with 70% ethanol and resuspended in DEPC-treated water. The digested RNA was used for cDNA synthesis and PCR amplification.
Mitochondrial Isolation
HFK698 and 18Nco cells were cultured in T75 flasks as described before. The cells were trypsinized, and about 5 × 108 cells were recovered by centrifugation at 600 × g for 10 min at 4 °C. The cells were washed with PBS and collected by centrifugation at 600 × g as described above. This procedure was repeated once. The final pellet was resuspended in 4 ml of a hypotonic solution containing 0.6 m mannitol, 1 mm EDTA, and 10 mm Hepes, pH 6.8, and incubated for 10 min on ice. The cells were homogenized by passing the suspension 15 times through a syringe coupled with a 23-gauge needle. The homogenization was monitored by phase microscopy until ∼70% of the cells were broken. The homogenate was centrifuged at 1,500 × g for 5 min at 4 °C, and the supernatant was recovered and centrifuged again as described above. The final supernatant was recovered and centrifuged at 10,000 × g for 30 min at 4 °C (9, 10, 19, 20). The final mitochondria pellet was resuspended in 2–3 ml of 0.25 m sucrose, 2 mm MgCl2, and 0.4 mm sodium phosphate buffer at pH 6.8 and treated with RNase A at a final concentration of 50 μg/ml for 15 min at room temperature (9, 10, 19, 20). The mitochondria fraction was recovered by centrifugation at 10,000 × g for 30 min and suspended in 100 μl of PBS containing 100 units of RNaseOut (Invitrogen), and mitochondrial RNA was extracted with TRIzol as described before. RT-PCR was carried out as described before using primers P12 (r) and P3 (f) for the SncmtRNA-1 and primers P13 (r) and P3 (f) for the SncmtRNA-2. Primers used to amplify mitochondrial COX I mRNA were as follows: 5′-TTCCGAAGCCTGGTAGGATAAGA (f) and 5′-GAACAGGTTGAACAGTCTACCCT (r). The 18S rRNA was used as a cytoplasmic transcript and was amplified using primers 5′-GTAACCCGTTGAACCCCATT (f) and 5′-CATCCAATCGGTAGTAGCGC (r).
In Situ Hybridization (ISH)
Cells cultured for 24 h in 8-well chamber slides (Lab-Tek, NUNC), were washed in PBS and fixed in 4% p-formaldehyde in PBS for 10 min at room temperature. The slides were then washed three times with PBS for 5 min and incubated with 0.2 n HCl for 10 min at room temperature. Hybridization was carried out essentially as described before (9, 10). For fluorescent in situ hybridization, after fixation, cells were hybridized for 18 h at 37 °C with 200 μl of the hybridization solution containing 3.5 pmol of the antisense probe (primer P8) or the corresponding sense probe (primer P9), previously labeled at the 3′-end with digoxigenin-11-dUTP (Roche Applied Science). The slides were washed with 2× SSC and 1× SSC for 10 min each at room temperature, 0.2× SSC for 30 min at 37 °C, and, finally, with 0.2× SSC for 10 min at room temperature. Cells were then incubated for 2 h at room temperature with anti-digoxigenin conjugated to fluorescein (Roche Applied Science), diluted 1:250 in blocking buffer (1% BSA, 0.3% Triton X-100 in PBS). The slides were washed in PBS for 10 min and then incubated for 15 min with DAPI solution (DAPI/PBS, 1:2000). Samples were mounted in Entellan (Merck) or Faramount (DAKO) and analyzed and photographed using Q-capturePro software in an Olympus BX-51 microscope.
Knockdown of SncmtRNA-1
SiHa or HeLa cells were plated onto 12-well plates (Nunc) at 2,5 × 104 cells/well. At 24 h, cells were transfected with 100 nm specific antisense oligonucleotide (ASO) (ASO-1 AS, 5′-GGTTTGGGGCTAGGTTTAGC) or control ASO (ASO-C, 5′-TTATATTTGTGTAGGGCTAG) (both ASOs full-phosphorothioate), using Lipofectamine 2000 (Invitrogen), according to the manufacturer's directions, or left untreated and incubated for 12, 24, or 48 h. At the indicated times, the cells were harvested and counted in quadruplicate in a Neubauer chamber under phase microscopy. These studies were carried out in triplicate. In order to determine the DNA synthesis rate, 2.5 × 104 SiHa or HeLa cells/well were plated onto 12-well plates (Nunc). The next day, cells were transfected with 100 nm ASO-1 AS or ASO-C using Lipofectamine 2000 (Invitrogen) or non-treated. Determination of DNA synthesis rate was carried out using the commercial kit Click-iT® EdU Alexa Fluor® 488 kit (Molecular Probes), according to manufacturer's directions. Briefly, 48 h after transfection, cells were pulsed with the nucleoside analog 5-ethynyl-2′-deoxyuridine (EdU; 10 μm) for 2 h at 37 °C. Cells were harvested, fixed in 3.7% p-formaldehyde in PBS, and saponin-permeabilized. EdU was detected and co-stained with DAPI. The cells were analyzed by fluorescence microscopy in an Olympus BX-51 microscope. In each experiment, the number of EdU-positive cells was counted versus the total amount of cells (DAPI staining) in at least quadruplicate.
RNA Interference of HPV-16 E4
Lentiviral vectors were constructed to generate short hairpin RNAs (shRNAs) targeted to HPV-16 E4, plus a control (siLuc) vector. Inverted self-complementary hairpin DNA oligonucleotides (Invitrogen) were ligated in a third generation self-inactivating lentiviral vector (pLL3.7) containing a CMV-driven EGFP reporter and a U6 promoter upstream from cloning restriction sites (HpaI and XhoI) to allow the introduction of oligonucleotides encoding shRNAs (21). Each hairpin consisted of a TG, a 19-nt sense sequence, a short spacer (TTCAAGAGA), the antisense sequence, 6 Ts (stop signal for RNA polymerase III), and an XhoI site. Oligonucleotides were annealed and inserted between the HpaI and XhoI sites of the plasmid. Primer pairs for constructing shE4 vectors were as follows: shE4B (f), 5′-TGCCAATCCTCACTGCATTTATTCAAGAGATAAATGCAGTGAGGATTGGCTTTTTTC; shE4B (r), 5′TCGAGAAAAAAGCCAATCCTCACTGCATTTATCTCTTGAATAAATGCAGTGAGGATTGGCA. Control shRNAs were as follows: shLUC (f), 5′TGTTCTCCGAACGTGTCACGTTTCAAGAGAACGTGACACGTTCGGAGAACTTTTTTC;shLUC reverse, 5′-TCGAAAAAAAGTTCTCCGAACGTGTCACGTTCTCTTGAAACGTGACACGTTCGGAGAACA.Restriction mapping and direct DNA sequencing confirmed correct insertions of shRNA cassettes. The constructed shRNA expression lentiviral vectors were named E4shRNA and ConshRNA.
Lentiviral vector production was carried out with 293FT cells essentially as described before (21). The cells were transfected with 9 μg of ViraPower® packaging mix and 3 μg of pLL3.7-E4B plasmid or control shRNA plasmid using 36 μl of Lipofectamine 2000 reagent (Invitrogen), according to the manufacturer's instructions. Forty-eight hours after co-transfection, viral particles were recovered and titrated using 293FT cells (21). Viral titers were in the range of 107 transducing units/ml of medium. To generate stable HFK698 lines, cells were cultured at 5 × 104 cells/well in 12-well plates along with recombinant lentivirus encoding shRNAs against E4 at a multiplicity of infection of 5 in keratinocyte serum-free medium/DMEM10 (3:1) medium at 37 °C and 5% CO2. At 72 h post-transduction, reporter gene expression (GFP) was examined using fluorescence microscopy. Two weeks following expansion, cells were then harvested by trypsin/EDTA, and fluorescent cells were sorted on a BDFACSAria II cell sorter. Gating was performed on the brightest cells, which were collected and subsequently expanded.
Expression of HPV-18 E1^E4 in HFK Cultures
Recombinant pCDNA3.1+Hyg vector, either empty or containing HPV-18 E1^E4 wild-type sequences, has been described elsewhere (22). Low passage pooled neonatal foreskin keratinocytes (Lonza, CH) were grown in serum-free medium (Invitrogen). Cells were transfected (Lipofectamine 2000) with recombinant plasmid expressing the neomycin resistance marker. After 24 h, the cells were selected with 20 μg/ml hygromycin B for 2 days to eliminate non-infected cells. Surviving cells were amplified and used to seed monolayer cultures.
Knockdown of HPV-16/18 E2
HFK698, HF18, and 18Nco cells were cultured in 12-well plates (Lab-Tek, NUNC) for 24 h and then washed with PBS. To each well, 900 μl of KSFM/DMEM10 (3:1) was added, and the cells were transfected with 100 nm full-phosphorothioate ASO directed against E2 (E2-ASO) from HPV-16 or -18 (23). The ASO was mixed with Lipofectamine 2000, according to the manufacturer's instructions (Invitrogen), in a final volume of 100 μl. After 20 min at room temperature, the 100-μl mix was added to the wells. Sequences of the ASOs used to knock down E2 HPV-16 and HPV-18 were as follows: 5′-CAATAATAGTCAACTTGACC and 5′-AAGCAGTGAGTAGGTTCTGTAT, respectively. The sequence of the control ASO (ASO-C) was 5′-GGCTACGTCCAGGAGCGCA-3′. Knockdown of E2 was evidenced by RT-PCR amplification from total RNA using primers specific for HPV-16 or -18 E2.
Statistical Analysis
To analyze the significance of each corresponding group of experiments, Student's test was used. Significance (p value) was set at the nominal level of p < 0.05 or less.
RESULTS
ASncmtRNAs Are Down-regulated in HPV-immortalized Cells
As seen in Fig. 1, and in accordance with our previous results (9, 10), normal HFK express the SncmtRNA-1 and ASncmtRNAs. In contrast, SiHa and HeLa cells, cervical cancer cell lines that arose from natural infection by HPV-16 and 18, respectively, express the SncmtRNA-1 and down-regulate expression of the ASncmtRNAs (Fig. 1A). Similarly, HFK immortalized with the complete genome of HPV-16 (HFK698 cells) (14) or HPV-18 (18Nco cells) (15) also express the SncmtRNA-1 and down-regulate the ASncmtRNAs (Fig. 1A).
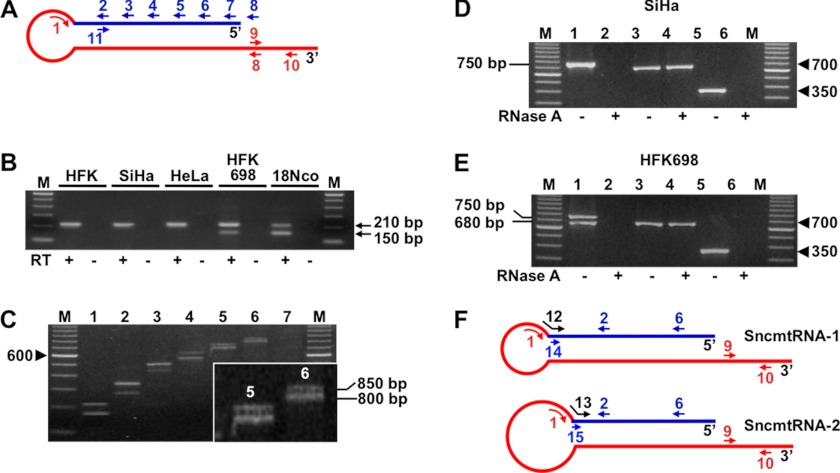
FIGURE 1.
Immortalization of HFK with HPV induces down-regulation of the ASncmtRNAs. A, expression of the SncmtRNA and the ASncmtRNAs in normal HFK or HFK immortalized with HPV-16 (HFK698 cells) or 18 (18Nco cells) was analyzed by ISH. The ASncmtRNAs are down-regulated in the immortalized cells as well as in the corresponding tumorigenic cell lines SiHa (HPV-16-transformed) and HeLa (HPV-18-transformed). Magnification was ×40. B, expression of SncmtRNA-1 (S), ASncmtRNA-1 (AS-1), and ASncmtRNA-2 (AS-2) in the different cell lines was evaluated by RT-PCR amplification. The internal control used was 18S rRNA. M, 100-bp ladder. C, the relative intensity of the bands corresponding to the different non-coding mitochondrial RNAs was determined by densitometry and normalized against the intensity of the 150-bp amplicon from 18S rRNA.
The differential expression of these mitochondrial transcripts assessed by ISH was confirmed by RT-PCR (see “Experimental Procedures”). The SncmtRNA-1 was amplified between the loop and the IR, between primers 1 and 2 (see Fig. 3A), to generate a fragment of 210 bp (9, 10) (Fig. 1B, S). The density of each band was analyzed by densitometry measurements (see “Experimental Procedures”). The 210-bp amplicon was normalized to the 150-bp amplicon from 18S rRNA, used as a loading control (Fig. 1B, 18S). The normalized amplicon of the SncmtRNA-1 indicates that the level of this transcript was similar in the five cell lines studied (Fig. 1C). Notice that only the RNA of HFK698 and 18Nco cells yielded an amplicon of about 150 bp with the same primers (Fig. 1B, S).
FIGURE 3.
HPV-16- or HPV-18-immortalized cells express a novel sense transcript or SncmtRNA-2. A, schematic structure of the SncmtRNA-1 and strategy to amplify the transcript between the loop and the IR. Red line, 16S mitochondrial rRNA; blue line, IR. The primers used for amplification are indicated. B, PCR amplification of cDNA from the indicated cells, using primers 1 and 2. The 210-bp fragment corresponds to the SncmtRNA-1, whereas the 150-bp fragment corresponds to the SncmtRNA-2. Lanes labeled 698 correspond to HFK698 cells. C, PCR amplification of HFK698 cDNA between primer 1 and primers 2, 3, 4, 5, 6, 7, and 8 (lanes 1–7, respectively). No amplification was observed when using primers 1 and 8 (lane 7). The inset shows the amplification fragments obtained with HFK698 cDNA and corresponding to lanes 5 and 6. M, 100-bp ladder. D and E, SncmtRNA-1 and SncmtRNA-2 contain a double-stranded region. D, RNA from SiHa cells in 2× SCC was incubated in the presence (even lanes) or absence (odd lanes) of 50 μg/ml RNase A for 15 s at room temperature. RNA was recovered and amplified by RT-PCR using primers 1 and 6 (lanes 1 and 2), primers 11 and 6 covering the double-stranded region (lanes 3 and 4), and primers 9 and 10 (lanes 5 and 6) targeted to the 3′ single-stranded region (see A). Only the double-stranded region was resistant to RNase digestion (lanes 3 and 4). E, similar results were obtained with total RNA from HFK698 cells. Amplification of the double amplicons with primers 1 and 6 (lanes 1 and 2) was abolished after digestion, and a similar result was obtained with primers 9 and 10 (lanes 5 and 6). These results indicate that the single-stranded regions (the loop and the 3′ region) of the SncmtRNA-2 were digested by RNase A. Amplification between primers 11 and 6, covering the double-stranded region of the SncmtRNA-2, was not affected by the enzyme (lanes 3 and 4). M, 100-bp ladder. F, schematic structure of the SncmtRNA-1 and SncmtRNA-2, indicating the primers used for amplification. Oligonucleotides 12 and 13 were used for specific RT-PCR amplification or for ISH. Primers 14 and 15 were used as controls.
A similar study was carried out with the antisense transcripts that were amplified also between the loop and the corresponding IR. As seen in Fig. 1B (AS-1), an amplicon of 220 bp corresponding to ASncmtRNA-1 was obtained after a 30-cycle amplification using HFK RNA. Only a weak band was obtained from total RNA obtained from the immortalized or the transformed cell lines. Normalization of the 220-bp amplicon against 18S rRNA revealed that the expression of ASncmtRNA-1 is inhibited in HFK698, 18Nco, SiHa, and HeLa cell lines (Fig. 1C). Similarly, the expression of ASncmtRNA-2 is down-regulated in HFK698, 18Nco, SiHa, and HeLa cells as compared with normal HFK (Fig. 1, B (AS-2) and C). Taken together, these results indicate that inhibition of the expression of ASncmtRNAs takes place during HPV-induced immortalization and before cellular transformation.
SncmtRNA-1 and Cell Proliferation
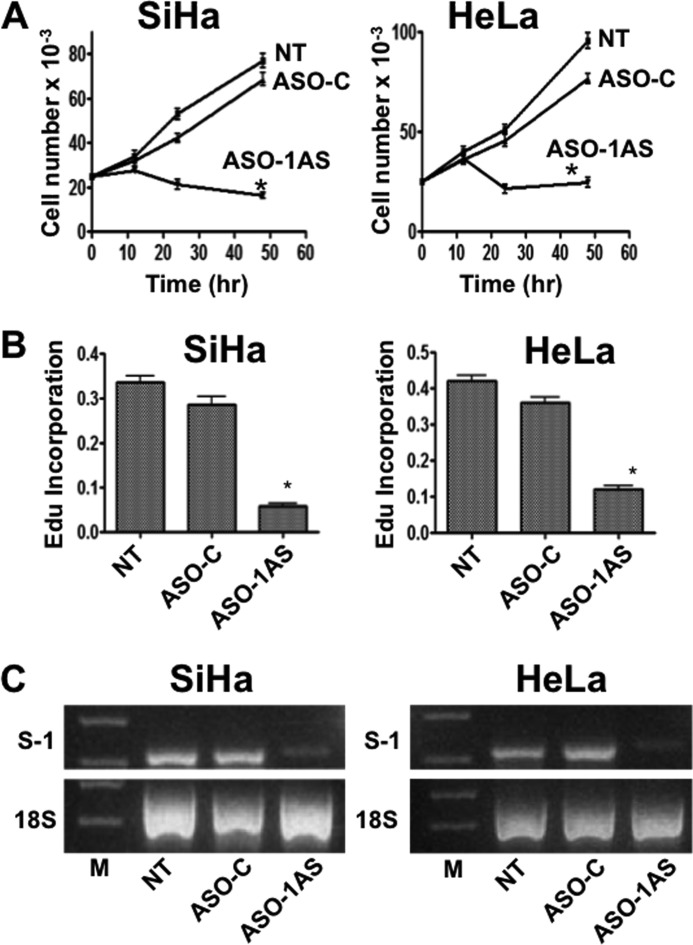
The close correlation between the expression of SncmtRNA-1 and the proliferative rate suggests a role for this mitochondrial transcript in regulation of the cell cycle. Therefore, we asked whether knockdown of SncmtRNA-1 would affect HeLa and SiHa proliferation. Knockdown of SncmtRNA-1 in SiHa and HeLa cells was induced with an antisense oligonucleotide (ASO-1AS) complementary to the loop region of the transcript (see “Experimental Procedures”). 25,000 cells/well were seeded onto 24-well plates and cultured overnight. The next day, the cells were transfected with 100 nm ASO-1AS or with ASO-C, using Lipofectamine2000. The cell number was determined in triplicate in three different assays at 12, 24, and 48 h post-transfection. Compared with non-transfected cells (NT) or cells transfected with ASO-C, knockdown of the SncmtRNA-1 induced a marked inhibition of proliferation in SiHa and HeLa cells (Fig. 2A). Similar results were obtained measuring the rate of DNA synthesis. As described, SiHa and HeLa cells were transfected with ASO-1AS or ASO-C for 48 h followed by a 2-h pulse with a BrdU analog (EdU, Molecular Probes). Knockdown of SncmtRNA-1 with 100 nm ASO-1AS inhibited DNA synthesis in SiHa and HeLa cells compared with untreated cells or cells transfected with ASO-C (Fig. 2B). Amplification of RNA from SiHa and HeLa cells by RT-PCR revealed that the SncmtRNA-1 was knocked down only after transfection for 48 h with ASO-1AS but not with ASO-C (Fig. 2C).
FIGURE 2.
Knockdown of SncmtRNA-1 induces inhibition of proliferation in SiHa and HeLa cells. A, cell proliferation. SiHa and HeLa cells (2.5 × 105 cells/well) were plated onto 12-well plates and the next day were transfected with 100 nm ASO-1 AS or ASO-Control (ASO-C) using Lipofectamine2000, or left untreated (NT). At the indicated time periods, cells were harvested and counted. The experiments were carried out in triplicate, and the vertical bars show the mean ± S.D. The asterisk at 48 h post-transfection indicates p < 0.05. B, DNA synthesis rate was determined with the Click-iT® EdU Alexa Fluor® 488 kit (Molecular Probes), according to the manufacturer's directions. After 48 h of transfection as described in A, cells were pulsed with 10 μm EdU for 2 h at 37 °C, harvested, fixed in 3.7% p-formaldehyde in PBS, and saponin-permeabilized. EdU was detected with and co-stained with DAPI. The cells were analyzed by fluorescence microscopy in an Olympus BX-51 microscope. For each experiment, the number of EdU-positive cells was counted in at least quadruplicate versus the total amount of cells (DAPI staining). The results are presented as the mean ± S.D. The asterisk on the ASO-1AS bars indicates p < 0.01 (SiHa) and p < 0.05 (HeLa), respectively. C, knockdown of the SncmtRNA-1 was confirmed by RT-PCR on total RNA extracted from SiHa and HeLa cells treated as in A. A representative gel from the experiment run in triplicate is shown. Transfection with ASO-1AS induces a marked reduction of the SncmtRNA-1 (S-1) as compared with the controls. The amplification of 18S rRNA (18S) used as loading control was not affected by the same treatment. M, 100-bp ladder.
Characterization of New Sense Transcript
As shown before (Fig. 1B), PCR amplification of SncmtRNA-1 from HFK, SiHa, HeLa, HFK698, and 18Nco cells using primer 1, targeted to the loop (Fig. 3A, red line) and primer 2, targeted to the IR (Fig. 3A, blue line), yielded the expected amplicon of around 210 bp (Fig. 3B) (9). Surprisingly, however, a second amplicon of about 150 bp was obtained only after amplification of cDNA from HFK698 and 18Nco cells (Fig. 1B, upper panel). The sequence of the 150-bp amplicon revealed a 120-nt IR linked to the 5′-end of the mitochondrial 16S ribosomal RNA (16S mtrRNA). To determine whether the IR of this new transcript, which we termed SncmtRNA-2, was longer than 120 nt, we used a PCR-walking strategy (9, 10). The cDNA from SiHa or HFK698 cells was amplified between the reverse primer 1 and the forward primers 2, 3, 4, 5, 6, 7, and 8, targeted to the putatively longer IR (Fig. 1B, blue line). In contrast to the single ladder of amplicons obtained with cDNA from SiHa cells (see supplemental Fig. 1), HFK698 cDNA yielded a ladder of double amplicons with the same set of primers (Fig. 1C). No amplification product was obtained using primers 1 and 8 (Fig. 1C, lane 7). The same results were obtained with cDNA from 18Nco cells (data not shown). The two amplicons of about 800 and 850 bp (Fig. 1C, lane 6, see inset) obtained with cDNA from HFK698 cells using primers 1 and 7, were separated by gel electrophoresis, purified, and sequenced. The sequence of the 800-bp amplicon revealed a 752-nt IR linked to the 5′-end of the 16S mtrRNA (see supplemental Fig. 2 and GenBankTM accession number HM581520). Compared with SncmtRNA-1, the IR of SncmtRNA-2 is 63 nt shorter at its 3′-end.
The presence of a 752-nt IR in the SncmtRNA-2 suggests that the transcript contains a long double-stranded region. To test this possibility, RNA from SiHa or HFK698 cells in 2× SCC was incubated in the presence (even lanes) or absence (odd lanes) of 50 μg/ml RNase A for 15 s at room temperature. RNA was recovered and amplified by RT-PCR using primers 1 and 6 (Fig. 3D, lanes 1 and 2), primers 11 and 6 covering the putative double-stranded region (Fig. 3D, lanes 3 and 4), and primers 9 and 10 (Fig. 3D, lanes 5 and 6) targeted to the 3′ single-stranded region (see Fig. 3A). Amplification was obtained after RNase digestion only with primers 11 and 6, confirming the double-stranded structure in this region (Fig. 3D, lanes 3 and 4). Similar results were obtained with total RNA from HFK698 cells. Amplification of the double amplicons with primers 1 and 6 (Fig. 3E, lanes 1 and 2) was abolished after digestion, and a similar result was obtained with primers 9 and 10 (Fig. 2E, lanes 5 and 6), indicating that the single-stranded regions (the loop and the 3′ region) of the SncmtRNA-2 were digested by RNase A. Amplification between primers 11 and 6, covering the double-stranded region of the transcript, was not affected by the enzyme (lanes 3 and 4). A deduced schematic representation of both transcripts is shown in Fig. 2F. Similarly to SncmtRNA-1 (9), SncmtRNA-2 also contains a stem-loop structure with a long double-stranded region (Fig. 2F).
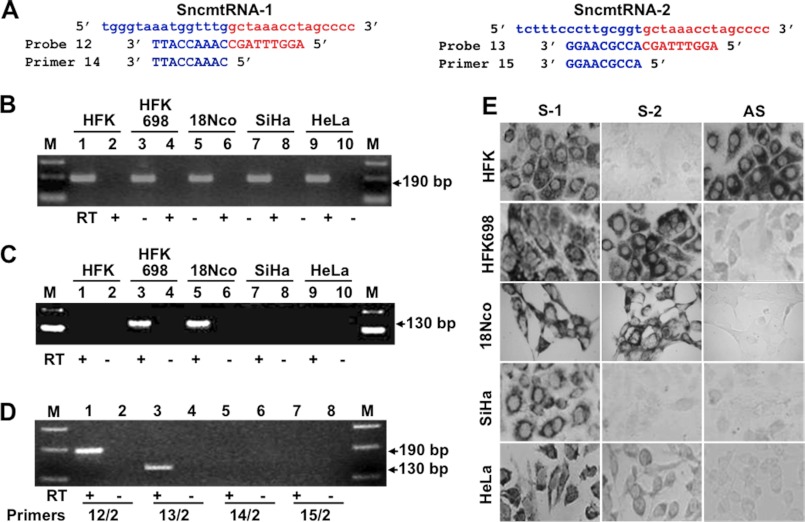
Expression of SncmtRNA-2
The ISH shown in Fig. 1A was carried out with probe 8 (Fig. 3A), which hybridizes to SncmtRNA-1 and SncmtRNA-2. To differentiate between these two transcripts, probes targeted to the linker region between the IR of each transcript and the 5′-end of the 16S mtrRNA were used. Probes 12 and 13 (Fig. 2F), specific for SncmtRNA-1 and SncmtRNA-2, respectively, contain 9 nt complementary to the 3′-end of the IR of each transcript followed by 9 nt targeted to the first 9 positions of the 16S mtrRNA (Fig. 4A). To test the specificity of these probes, cDNA from HFK, HFK698, 18Nco, SiHa, and HeLa cells was amplified using primers 12 and 2, resulting in a 190-bp amplicon (Fig. 4B), which corresponds to SncmtRNA-1. Amplification with primers 13 and 2 yielded an amplicon of about 130 bp only with cDNA from HFK698 and 18Nco cells (Fig. 4C). The sequence of this amplicon confirmed the structure of the SncmtRNA-2 (data not shown). As expected, no amplification was obtained with primer 2 and a reverse primer of 9 nt (Fig. 3F, primer 14) complementary to the 3′-end of the IR of the SncmtRNA-1 (Fig. 4D, lanes 5 and 6) or the SncmtRNA-2 (primer 15) (Fig. 4D, lanes 7 and 8). Therefore, probes 12 and 13 were used to distinguish SncmtRNA-1 and SncmtRNA-2 by ISH. As shown in Fig. 4E, ISH with probe 12 revealed the expression of SncmtRNA-1 (S-1) in HFK, HFK698, 18Nco, SiHa, and HeLa cells. On the other hand, hybridization with probe 13 revealed the expression of SncmtRNA-2, only in HFK698 and 18Nco cells (Fig. 4E, S-2). The ASncmtRNAs were expressed only in HFK (Fig. 4E, AS).
FIGURE 4.
Specific detection of SncmtRNA-1 and SncmtRNA-2. A, sequence of probes 12 (specific for SncmtRNA-1) and 13 (specific for SncmtRNA-2) targeted to the linker region between the IR and the 5′-end of the 16S mtrRNA (see Fig. 3F, showing the position of these primers and control primers 14 and 15). B, PCR amplification of cDNA obtained from the indicated cells using primers 12 and 2 (SncmtRNA-1; Fig. 3F). A single fragment of 190 bp was obtained in all samples. C, same as B but using primers 13 and 2 (SncmtRNA-2; Fig. 3F). A 130-bp fragment corresponding to SncmtRNA-2 was obtained only with cDNA from HFK698 and 18Nco cells. M, 100-bp ladder. D, PCR amplification of SncmtRNA-1 and -2 from HFK698 cDNA. The expected fragments of 190 and 130 bp were obtained using primer 2 in combination with primers 12 or 13 (lanes 1–4). No amplification was observed when primer 2 was used in combination with primer 14 (9 nt complementary to the 5′-end of the 16S rRNA) or primer 15 (9 nt complementary to the 3′-end of the IR) of the SncmtRNA-1. Lane M, 100-bp ladder. E, differential expression of the SncmtRNA-1 and SncmtRNA-2. The expression of SncmtRNA-1 (S-1) and SncmtRNA-2 (S-2) in the indicated cells was determined by ISH using probe 12 and probe 13, respectively. The SncmtRNA-2 was expressed only in the immortalized cells HFK698 and 18Nco. Notice that only HFK expresses the ASncmtRNAs (AS). Magnification was ×40.
Extramitochondrial Localization of SncmtRNA-2
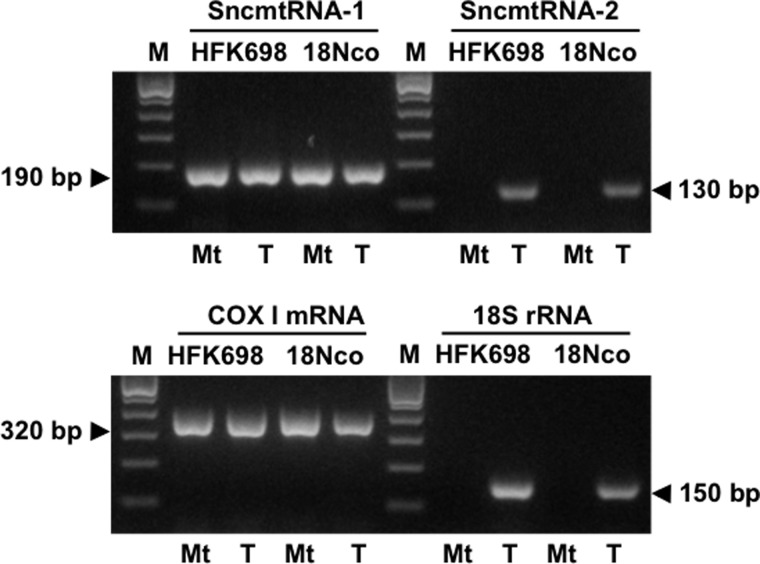
We reported before that the SncmtRNA-1 and the ASncmtRNAs were localized in mitochondria isolated from HFK cells and mitogen-activated human lymphocytes (10). SncmtRNA-2 exhibits 99,7% identity with the sequence of the human 16S mitochondrial gene, and therefore one would expect that this transcript is also found localized in the organelle. Mitochondria were isolated from HFK698 and 18Nco cells and treated externally with RNase A to eliminate cytoplasmic RNA contamination (9, 10, 19, 20). RNA was extracted from the isolated mitochondria and amplified by RT-PCR. To ascertain that RNase treatment eliminated cytoplasmic RNA contamination, total RNA was also extracted from HFK698 and 18Nco cells. Amplification was carried out using primer pairs for SncmtRNA-1, SncmtRNA-2, COX I, and 18S rRNA (see “Experimental Procedures”). SncmtRNA-1 and COX I mRNA were readily amplified from RNA of nuclease-treated mitochondria and total RNA from HFK698 and 18Nco cells (Fig. 5). As expected, the 18S rRNA was amplified from total RNA but not from nuclease-treated mitochondria (Fig. 5). Interestingly, and in contrast to SncmtRNA-1, SncmtRNA-2 was amplified from total RNA of HFK698 and 18Nco but not from mitochondrial RNA (Fig. 5, SncmtRNA-2). These results indicate that the SncmtRNA-2 is found only outside the organelle.
FIGURE 5.
The SncmtRNA-2 is not present in isolated mitochondria. Mitochondria were isolated from ∼5 × 108 HFK698 or 18Nco cells as described under “Experimental Procedures.” The final mitochondrial fraction was treated with RNase A to eliminate contamination from cytoplasmic RNA, followed by extraction of mitochondrial RNA with TRIzol. In parallel, total RNA was also extracted from HFK698 and 18Nco cells. Amplification of fragments of 190 bp (SncmtRNA-1), 130 bp (SncmtRNA-2), 320 bp (COX I mRNA), and 150 bp corresponding to 18S rRNA was observed in total RNA from both immortalized cell lines (T lanes). Only the amplicons of 190 bp of the SncmtRNA-1 and 320 bp of the COX I mRNA were amplified from mitochondrial RNA (Mt lanes) of HFK698 and 18Nco cells, whereas no amplification was obtained of the SncmtRNA-2 or 18S rRNA. M, 100-bp ladder.
HPV-16/18 Oncogenes Modulate Expression of Mitochondrial ncRNAs
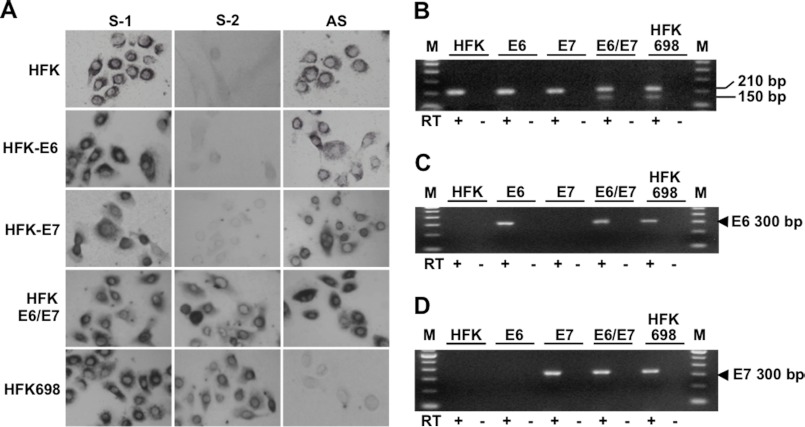
We next examined which oncogene(s) of HPV-16 is involved in down-regulation of the ASncmtRNAs and in the expression of the SncmtRNA-2. HFK were transduced with retroviral vectors containing the coding sequences of E6, E7, or E6 and E7 (16, 17). Probes 12 and 13 were used to determine the expression of SncmtRNA-1 and SncmtRNA-2, respectively, by ISH. HFK transduced with E6 or E7 express the SncmtRNA-1 (S-1) and the ASncmtRNAs (AS) (Fig. 6A), but not SncmtRNA-2 (Fig. 6A, S-2), just as normal keratinocytes. Interestingly, however, transduction of HFK with E6 and E7 induced expression of the SncmtRNA-2, just like the immortalized cell line HFK698 (Fig. 6A, S-2). RT-PCR amplification using primers 1 and 2 (see Fig. 3A) confirmed that the 150-bp fragment corresponding to SncmtRNA-2 was amplified only in HFK transduced with E6/E7 as well as in HFK698 cells (Fig. 6B). Expression of E6 and E7 mRNA was confirmed by RT-PCR amplification. E6 is expressed in HFK698 cells, or in HFK transduced with E6 or with E6/E7 (Fig. 6C), whereas E7 mRNA is expressed in HFK transduced with E7 or E6/E7 and in HFK698 cells (Fig. 6D).
FIGURE 6.
Expression of SncmtRNA-2 is induced by HPV-16 E6 and E7. A, expression of the SncmtRNA-1, SncmtRNA-2, and the ASncmtRNAs was determined in HFK transduced with E6, E7, or E6/E7. The SncmtRNA-1 was expressed in all cells, including HFK698. The SncmtRNA-2 was expressed only in HFK transduced with E6 and E7 and in HFK698. Expression of the ASncmtRNAs was maintained in all cells except HFK698. Magnification was ×20. B, PCR amplification confirmed that the SncmtRNA-2 was expressed only in HFK698 and HFK transduced with E6 and E7. C, total RNA from HFK698 cells, HFK, and HFK transduced with E6, E7, and E6/E7 was used for RT-PCR amplification of HPV-16 E6 mRNA. E6 is expressed in HFK transduced with E6 or E6/E7 and HFK698 cells. D, same as C, but amplification was carried out with primers corresponding to HPV-16 E7 mRNA. Lane M, 100-bp ladder.
E2 Oncogene from HPV-16 or -18 Induces Down-regulation of ASncmtRNAs
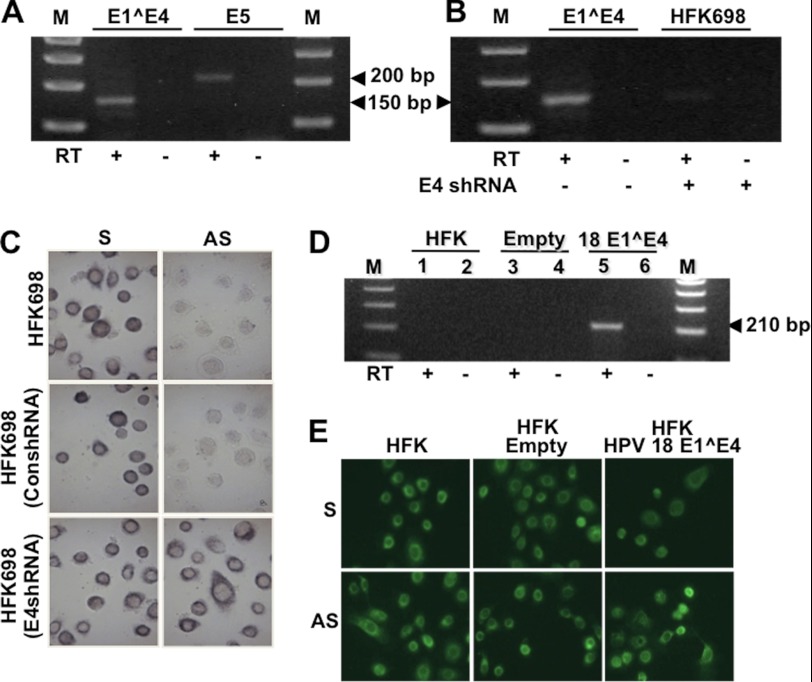
Because expression of the ASncmtRNAs is not affected by E6, E7, or E6 and E7 (Fig. 6A, AS), we asked whether other HPV-16 proteins expressed in HFK698 might be involved in down-regulation of these transcripts. A reasonable candidate is E1^E4 because this protein is known to interact with mitochondria (24) and is expressed in HFK698 cells together with E5 (Fig. 7A). To knock down E1^E4, HFK698 cells were transduced with a lentiviral vector that expresses GFP and an shRNA targeted to HPV-16 E1^E4 (21). The GFP-expressing HFK698 cells were sorted and then cultured for 24 h. As shown in Fig. 7B, the specific shRNA induced knockdown of E1^E4, and, concomitantly, expression of the ASncmtRNAs was reestablished (Fig. 7C, E4-shRNA). Notice that the control shRNA did not affect the expression of the ASncmtRNAs (Fig. 7C, ConshRNA). Using an inverse strategy, HFK cells were transfected with a plasmid containing HPV-18 E1^E4 (22). The transfected cells were selected with gentamycin and cultured for 24 h. RT-PCR confirmed that the transfected cells, but not HFK cells or HFK cells transfected with empty vector, express E1^E4 (Fig. 7D). However, the expression of the ASncmtRNAs was not affected by E1^E4 (Fig. 7E).
FIGURE 7.
Down-regulation of the ASncmtRNA and HPV oncogenes. A, HFK698 cells express E1^E4 and E5 mRNA. B, HFK698 cells were transduced with a GFP-expressing lentiviral vector encoding an shRNA complementary to E4. The GFP-expressing cells were sorted, and expression of E1^E4 mRNA was determined by RT-PCR amplification. E4 was knocked down in HFK698 cells transduced with the E4shRNA. C, expression of the SncmtRNA-1 and the ASncmtRNAs in HFK698 cells transduced with E4shRNA or empty vector (ConshRNA). Expression of the ASncmtRNAs was reestablished only in HFK698 cells transduced with the E4shRNA. Magnification was ×20. D, HFK cells were transformed with a plasmid containing HPV-18 E1^E4 or with empty vector. E1^E4 was expressed only in HFK transformed with E1^E4-containing vector. E, HFK cells transformed with HPV-18 E1^E4 plasmid or with the empty vector were subjected to fluorescent in situ hybridization and compared with untreated HFK. The expression of the SncmtRNA-1 and the ASncmtRNAs was not affected by the expression of E1^E4. Magnification was ×20.
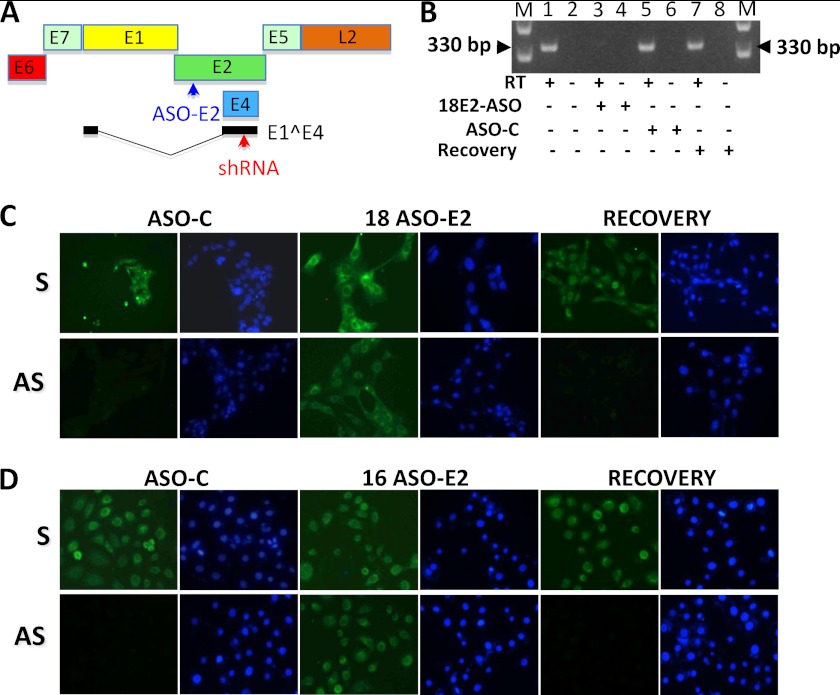
E1^E4 results from a transplicing reaction between the first 15 nt of E1 and a fragment of 264 nt positioned close to the 3′-end of E2, as illustrated in Fig. 8A (25). Therefore, the E4-shRNA should also induce knockdown of E2 (Fig. 8A, shRNA, red arrow). To explore whether E2 is indeed involved in down-regulation of the ASncmtRNAs, 18Nco cells were transfected with an ASO complementary to the NH2-terminal region of HPV-18 E2 (Fig. 6A, E2-ASO, blue arrow) or with ASO-C. Twenty-four hours after transfection, cells were used to prepare total RNA or fixed to determine the expression of the ASncmtRNAs. As shown in Fig. 8B, E2-ASO induced knockdown of E2 in 18Nco cells, whereas the control ASO had no effect (Fig. 8B, C-ASO). Interestingly, fluorescent in situ hybridization revealed that the expression of the ASncmtRNAs was reestablished in 18Nco cells upon knockdown of E2 (Fig. 8C, 18 E2-ASO). Transfection with ASO-C had no effect (Fig. 8C, C-ASO). To determine whether the effect on the expression of the ASncmtRNAs was reversible, E2-ASO was removed, and the cells were further cultured in normal medium for an additional 24 h. Together with the recovery of E2 expression (Fig. 6B, Recovery), the ASncmtRNAs were again down-regulated (Fig. 8C, Recovery). The same results were obtained with HFK698 (Fig. 8D and supplemental Fig. 2A) and HF18 cells (keratinocytes immortalized with HPV-18) (see supplemental Fig. 3, B and C). Transfection of 18Nco or HFK698 cell lines with E2-ASO did not affect the expression of E6, E7, E1, and E5 (see supplemental Fig. 4, A and B).
FIGURE 8.
HPV E2 is involved in down-regulation of the ASncmtRNAs. A, schematic illustration of the HPV-16 genome, indicating the position of E6, E7, E1, E2, E4, E5, and L2 genes. The absence of L1 is for simplicity. The transplicing reaction between a fragment of E1 with a fragment of E2 to generate E1^E4 is shown. B, expression of E2 mRNA in 18Nco cells was determined by RT-PCR amplification. A fragment of 330 bp was obtained with cDNA of 18Nco cells (lanes 1 and 2). E2 was knocked down by an ASO complementary to the NH2-terminal coding region (18 E2-ASO; lanes 3 and 4) but not by a control oligonucleotide (C-ASO; lanes 5 and 6). 24 h after transfection with the 18E2-ASO, the cells were cultured in fresh medium for another 24 h. The expression of E2 was reestablished (Recovery; lanes 7 and 8). C, analysis of the expression of SncmtRNA-1 and -2 (S) and ASncmtRNAs (AS) by fluorescent in situ hybridization. Hybridization revealed that the expression of the ASncmtRNAs was recovered in 18Nco cells transfected with the 18E2-ASO. Once the expression of E2 was reestablished, expression of the ASncmtRNAs was down-regulated (Recovery). D, the same results were obtained with HFK698 cells transfected with an E2-ASO complementary to HPV-16 E2 (16 E2-ASO). The expression of the ASncmtRNAs was reestablished after knocking down HPV-16 E2. Recovery of E2 expression resulted in down-regulation of the ASncmtRNAs (Recovery). Magnification was ×20.
DISCUSSION
The function of the ASncmtRNAs is unclear. Interestingly, however, the ASncmtRNAs are down-regulated in several tumor cell lines as well as in tumor cells present in biopsies of 17 different types of cancer and patients (10). A hallmark of cancer is the inactivation of tumor suppressors together with constitutive activation of oncogenes, resistance to apoptosis, and metastasis (1). Therefore, down-regulation of the ASncmtRNAs suggests, hypothetically, that these transcripts might function as a novel mitochondrially encoded tumor suppressor (10). Interestingly, nuclear encoded ncRNAs function as either oncogenes or tumor suppressors (26–28). In general, the evidence to assign the tumor suppressor tag relates to down-regulation of these molecules in cancer cells as compared with normal counterparts (29). However, the designation of tumor suppressor requires experimental evidence showing the role of this class of molecules in cell cycle control.
Indeed, the aim of this work was to shed some light on the mechanisms involved in down-regulation of the ASncmtRNAs during early carcinogenesis. To achieve this goal, we used as a model HFK immortalized with high risk HPV. The results indicate that E2 is involved in down-regulation of the ASncmtRNAs in HFK immortalized with high risk HPV-16 and HPV-18. E2 is essential for viral genome replication and regulation of E6 and E7 expression in early stages after HPV infection (5, 6, 30–32). In addition, regulation of transcription factors, cell proliferation, apoptosis, cell differentiation, and chromosome instability seem to be the most significant functions of E2 (33–40). Interestingly, and in relation to the present work, E2 seems to have oncogenic potential by itself. Expression of the HPV-8 E2 gene in transgenic mice results in increased skin cancer development, which is enhanced by UV irradiation (41, 42). On the other hand, HPV-11 E2, a low risk HPV virus, does not induce cell transformation (43). Whether the induction of skin cancer is related to the ability of HPV-8 E2 to induce knockdown of the ASncmtRNAs warrants future research.
However, the reversible effect of E2 on the expression of the ASncmtRNAs raises a paradox. Although SiHa and HeLa cells do not express E2 (see supplemental Fig. 5), the ASncmtRNAs are down-regulated. One possibility is that after transformation into the tumorigenic lineage, another cellular protein could replace the ability of E2 to induce down-regulation of the ASncmtRNAs. Interestingly, the tumorigenic cell line TC-1, a mouse lung epithelial cell line immortalized with HPV-16 E6 and E7 and transformed with H-Ras oncogene (44), also exhibits down-regulation of the ASncmtRNAs. Here we show that HFK transduced with E6 and E7 do not affect the expression of the ASncmtRNAs (Fig. 6A). Therefore, down-regulation of these transcripts in TC-1 cells could be triggered by H-Ras, suggesting a role for this cellular oncogene similar to that of E2 in the modulation of the expression of ASncmtRNAs.4 Altogether, these results strongly suggest that an important cellular alteration occurring during HPV-induced carcinogenesis is inhibition of the expression of the ASncmtRNAs. One is tempted to speculate that some gene products of other oncogenic viruses might also induce down-regulation of the ASncmtRNAs (2–4). Interestingly, the ASncmtRNAs are down-regulated in HEK 293 cells (transformed with adenovirus) and the lymphoma cell line Devernelle (transformed with Epstein-Barr virus).4
Previous work has shown nucleo-cytoplasmic localization of high risk HPV-16/18 E2 (45). As far as we know, E2 does not interact with mitochondria, and therefore the pertinent question is how E2 alone or in combination with other cellular factors induces down-regulation of the ASncmtRNAs. Electron microscopy ISH showed that the SncmtRNA and the ASncmtRNAs in normal human kidney exit the organelle and are found localized in the cytoplasm and in the nucleus associated with chromatin and nucleoli (13). In renal cell carcinoma, the SncmtRNA shows similar localization, whereas the few copies of the ASncmtRNAs are mainly found in the cytoplasm (13). Therefore, an intriguing question is how, if so, these mitochondrial transcripts containing long double-stranded regions escape from the processing activities of Dicer and Drosha (46). One hypothetical possibility is that the double-stranded region of these mitochondrial RNA binds to the double-stranded binding domain of Dicer and/or Drosha (47, 48), resulting in inhibition of their activities. It has been demonstrated that RNAs with double-stranded structures inhibit Dicer or Drosha, such as ncRNAs from adenovirus (49). In Caenorhabditis elegans, the rncs-1 ncRNA contains a long double-stranded structure that inhibits Dicer (50). Perhaps then, the ASncmtRNAs in HFK are forming complexes with Dicer, inhibiting its processing activity. Therefore, the expression of E2 in HPV-immortalized HFK would relieve the inhibition on Dicer, resulting in degradation or processing of the double-stranded structure of the ASncmtRNAs.
Critical for HFK immortalization is the ability of the HPV oncogenes E6 and E7 to block the function of the tumor suppressors p53 and Rb together with up-regulation of telomerase (5, 6, 14, 15, 51–54). As reported here, an additional function of E6 and E7 is to induce the expression of the SncmtRNA-2 in primary keratinocytes. Why both oncoproteins are needed for the expression of SncmtRNA-2 is unclear, but it is interesting that efficient HFK immortalization as well as full up-regulation of telomerase also requires both E6 and E7 expression (54). As described before, the expression of the SncmtRNA-1 closely correlates with cell proliferation (9, 10). Here we show that knockdown of SncmtRNA-1 induces a marked inhibition of cell proliferation and DNA synthesis in SiHa and HeLa cells (see Fig. 2). These results indicate for the first time that SncmtRNA-1 is a new mitochondrial component participating in the regulation of the cell cycle. Hypothetically, the inhibition of proliferation of SiHa and HeLa cells might have a potential therapeutic application in cervical cancer.
Why immortalized cells require SncmtRNA-2 in addition to SncmtRNA-1 is unknown and warrants future investigation. We showed that the SncmtRNA-1 and the ASncmtRNAs are present in isolated mitochondria (9, 10). Here we confirmed that SncmtRNA-1 is present together with COX I mRNA in isolated mitochondria of the immortalized cell lines, but not SncmtRNA-2 (see Fig. 5). These results suggest that a fraction of SncmtRNA-1 is processed outside of the organelle, to give rise to SncmtRNA-2 and a fragment of 63 nt released from the IR. Interestingly, we have shown that the corresponding murine SncmtRNA-1 (named chimeric RNA) is also processed outside the mitochondria by an editing reaction from U to C (55). The nature of the processing mechanism of SncmtRNA-1 to SncmtRNA-2 plus the fragment of 63 nt is unclear. However, this reaction might be similar to the cleavage-and-ligation reactions (editosome) necessary for the edition of kinetoplastid transcripts in Trypanosoma and Leishmania (56–58). The question then is which molecule is important for HPV-immortalized cells: the SncmtRNA-2 or the 63-nt fragment released from the IR of the SncmtRNA-1? We are tempted to speculate that the important molecule is the fragment of 63 nt, based on in silico analysis of this sequence. Interestingly, these studies revealed that the 63-nt fragment was highly complementary only to microRNA-620 or hsa-miR-620 (Supplemental Fig. 6). Using a target scan algorithm, we found that hsa-miR-620 is involved in the silencing of more than 100 target mRNAs (68). An interesting example is the mRNA of promyelocytic leukemia (PML) protein, which is a core component of PML nuclear bodies found in tumor cells (59, 60). PML nuclear bodies are important structures involved in HPV replication, and several reports indicate that the E6 and E7 oncoproteins are localized in these nuclear structures (61–63). As shown here, E6 and E7 are needed for the expression of SncmtRNA-2, and therefore it is tempting to suggest that the 63-nt fragment released from SncmtRNA-1 is required to work as a “sponge” (64, 65) to trap hsa-miR-620 and consequently relieve the negative effect of this microRNA on the expression of PML mRNA. Interestingly, a ncRNA from herpesvirus saimirí binds and induces degradation of miR-27 to facilitate infection and viral transformation (66).
Finally, it is unclear why SncmtRNA-2 is not expressed in SiHa or HeLa cells, which constitutively express E6 and E7 (supplemental Fig. 5, A and B). Perhaps other cellular factors expressed after HPV transformation block the expression of SncmtRNA-2. Nevertheless, the fact that SncmtRNA-2 is expressed in immortalized but not tumorigenic cells might contribute to the screening of early cervical intraepithelial premalignant lesions (67).
This work was supported by Fondecyt Grants 11090060, 1085210, and 1110835; Fondef Grant D04I1338; CCTE-PFB16 Program Grant; Conicyt, Chile, and Universidad Andrés Bello, Chile, Grants DI-34-09/R, DI-31-09/R, DI-28-09/R4, and DI-06-09/R.

This article contains supplemental Figs. 1–6.
C. Villota, A. Campos, V. A. Burzio, M. Varas, and L. O. Burzio, unpublished data.
- HPV
- human papillomavirus
- ncRNA
- non-coding RNA
- SncmtRNA
- sense non-coding mitochondrial RNA
- ASncmtRNA
- antisense non-coding mitochondrial RNA
- HFK
- human foreskin keratinocytes
- ISH
- in situ hybridization
- ASO
- antisense oligodeoxynucleotide
- ASO-C
- control ASO
- IR
- inverted repeat
- nt
- nucleotide(s)
- EdU
- 5-ethynyl-2′-deoxyuridine
- 16S mtrRNA
- mitochondrial 16S ribosomal RNA
- PML
- promyelocytic leukemia.
REFERENCES
- 1. Hanahan D., Weinberg R. A. (2011) Hallmarks of cancer. The next generation. Cell 144, 646–674 [DOI] [PubMed] [Google Scholar]
- 2. D'Agostino D. M., Bernardi P., Chieco-Bianchi L., Ciminale V. (2005) Mitochondria as functional targets of proteins coded by human tumor viruses. Adv. Cancer Res. 94, 87–142 [DOI] [PubMed] [Google Scholar]
- 3. McLaughlin-Drubin M. E., Munger K. (2008) Viruses associated with human cancer. Biochim. Biophys. Acta 1782, 127–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McLaughlin-Drubin M. E., Münger K. (2009) Oncogenic activities of human papillomaviruses. Virus Res. 143, 195–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. zur Hausen H. (2009) Papillomaviruses in the causation of human cancers. A brief historical account. Virology 384, 260–265 [DOI] [PubMed] [Google Scholar]
- 6. zur Hausen H. (2002) Papillomaviruses and cancer. From basic studies to clinical application. Nat. Rev. Cancer 2, 342–350 [DOI] [PubMed] [Google Scholar]
- 7. Kroemer G. (2006) Mitochondria in cancer. Oncogene 25, 4630–4632 [DOI] [PubMed] [Google Scholar]
- 8. Chatterjee A., Dasgupta S., Sidransky D. (2011) Mitochondrial subversion in cancer. Cancer Prev. Res. 4, 638–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Villegas J., Burzio V., Villota C., Landerer E., Martinez R., Santander M., Martinez R., Pinto R., Vera M. I., Boccardo E., Villa L. L., Burzio L. O. (2007) Expression of a novel non-coding mitochondrial RNA in human proliferating cells. Nucleic Acids Res. 35, 7336–7347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Burzio V. A., Villota C., Villegas J., Landerer E., Boccardo E., Villa L. L., Martínez R., Lopez C., Gaete F., Toro V., Rodriguez X., Burzio L. O. (2009) Expression of a family of noncoding mitochondrial RNAs distinguishes normal from cancer cells. Proc. Natl. Acad. Sci. U.S.A. 106, 9430–9434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Felty Q., Singh K. P., Roy D. (2005) Estrogen-induced G1/S transition of G0-arrested estrogen-dependent breast cancer cells is regulated by mitochondrial oxidant signaling. Oncogene 24, 4883–4893 [DOI] [PubMed] [Google Scholar]
- 12. Sligh J. E., Levy S. E., Waymire K. G., Allard P., Dillehay D. L., Nusinowitz S., Heckenlively J. R., MacGregor G. R., Wallace D. C. (2000) Maternal germ line transmission of mutant mtDNAs from embryonic stem cell-derived chimeric mice. Proc. Natl. Acad. Sci. U.S.A. 97, 14461–14466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Landerer L., Villegas J., Burzio V. A., Oliveira L., Villota C., Lopez C., Restovic F., Martinez R., Castillo O., Burzio L. O. (2011) Nuclear localization of the mitochondrial ncRNAs in normal and cancer cells. Cell Oncol. 34, 297–305 [DOI] [PubMed] [Google Scholar]
- 14. Barbosa M. S., Schlegel R. (1989) The E6 and E7 genes of HPV-18 are sufficient for inducing two-stage in vitro transformation of human keratinocytes. Oncogene 4, 1529–1532 [PubMed] [Google Scholar]
- 15. Romanczuk H., Villa L. L., Schlegel R., Howley P. M. (1991) The viral transcriptional regulatory region upstream of the E6 and E7 genes is a major determinant of the differential immortalization activities of human papillomavirus types 16 and 18. J. Virol. 62, 2739–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Halbert C. L., Demers G. W., Galloway D. A. (1991) The E7 gene of human papillomavirus type 16 is sufficient for immortalization of human epithelial cells. J. Virol. 65, 473–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Boccardo E., Manzini Baldi C. V., Carvalho A. F., Rabachini T., Torres C., Barreta L. A., Brentani H., Villa L. L. (2010) Expression of human papillomavirus type 16 E7 oncoprotein alters keratinocytes expression profile in response to tumor necrosis factor-α. Carcinogenesis 31, 521–531 [DOI] [PubMed] [Google Scholar]
- 18. Young P. G., Attardi G. (1975) Characterization of double-stranded RNA from HeLa cell mitochondria. Biochem. Biophys. Res. Commun. 65, 1201–1207 [DOI] [PubMed] [Google Scholar]
- 19. Gaines G., Attardi G. (1984) Highly efficient RNA-synthesizing system that uses isolated human mitochondria. New initiation events and in vivo-like processing patterns. Mol. Cell Biol. 4, 1605–1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kruse B., Murdter N. N., Attardi G. (1995) Transcription system using a HeLa cell mitochondrial lysate. Methods Mol. Biol. 37, 179–197 [DOI] [PubMed] [Google Scholar]
- 21. Rubinson D. A., Dillon C. P., Kwiatkowski A. V., Sievers C., Yang L., Kopinja J., Rooney D. L., Zhang M., Ihrig M. M., McManus M. T., Gertler F. B., Scott M. L., Van Parijs L. (2003) A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat. Genet. 33, 401–406 [DOI] [PubMed] [Google Scholar]
- 22. Knight G. L., Pugh A. G., Yates E., Bell I., Wilson R., Moody C. A., Laimins L. A., Roberts S. (2011) A cyclin-binding motif in human papillomavirus type 18 (HPV18) E1^E4 is necessary for association with CDK-cyclin complexes and G2/M cell cycle arrest of keratinocytes but is not required for differentiation-dependent viral genome amplification or L1 capsid protein expression. Virology 412, 196–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Alvarez-Salas L. M., Benítez-Hess M. L., DiPaolo J. A. (2003) Advances in the development of ribozymes and antisense oligodeoxynucleotides as antiviral agents for human papillomaviruses. Antivir. Ther. 8, 265–278 [PubMed] [Google Scholar]
- 24. Raj K., Berguerand S., Southern S., Doorbar J., Beard P. (2004) E1 empty set E4 protein of human papillomavirus type 16 associates with mitochondria. J. Virol. 78, 7199–7207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Doorbar J., Ely S., Sterling J., McLean C., Crawford L. (1991) Specific interaction between HPV-16 E1–E4 and cytokeratins results in collapse of the epithelial cell intermediate filament network. Nature 352, 824–827 [DOI] [PubMed] [Google Scholar]
- 26. Mattick J. S., Makunin I. V. (2006) Non-coding RNA. Hum. Mol. Genet. 15, R17–R29 [DOI] [PubMed] [Google Scholar]
- 27. Esquela-Kerscher A., Slack F. J. (2006) Oncomirs. MicroRNAs with a role in cancer. Nat. Rev. Cancer 6, 259–269 [DOI] [PubMed] [Google Scholar]
- 28. Lee S. K., Calin G. A. (2011) Non-coding RNAs and cancer. New paradigms in oncology. Discov. Med. 11, 245–254 [PubMed] [Google Scholar]
- 29. Payne S. R., Kemp C. J. (2005) Tumor suppressor genetics. Carcinogenesis 26, 2031–2045 [DOI] [PubMed] [Google Scholar]
- 30. Bernard B. A., Bailly C., Lenoir M. C., Darmon M., Thierry F., Yaniv M. (1989) The human papillomavirus type 18 (HPV18) E2 gene product is a repressor of the HPV18 regulatory region in human keratinocytes. J. Virol. 63, 4317–4324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Romanczuk H., Thierry F., Howley P. M. (1990) Mutational analysis of cis elements involved in E2 modulation of human papillomavirus type 16 P97 and type 18 P105 promoters. J. Virol. 64, 2849–2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Steger G., Corbach S. (1997) Dose-dependent regulation of the early promoter of human papillomavirus type 18 by the viral E2 protein. J. Virol. 71, 50–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Smith J. A., White E. A., Sowa M. E., Powell M. L., Ottinger M., Harper J. W., Howley P. M. (2010) Genome-wide siRNA screen identifies SMCX, EP400, and Brd4 as E2-dependent regulators of human papillomavirus oncogene expression. Proc. Natl. Acad. Sci. U.S.A. 107, 3752–3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ramírez-Salazar E., Centeno F., Nieto K., Valencia-Hernández A., Salcedo M., Garrido E. (2011) HPV16 E2 could act as down-regulator in cellular genes implicated in apoptosis, proliferation, and cell differentiation. Virol. J. 8, 247–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee D., Kim H. Z., Jeong K. W., Shim Y. S., Horikawa I., Barrett J. C., Choe J. (2002) Human papillomavirus E2 down-regulates the human telomerase reverse transcriptase promoter. J. Biol. Chem. 277, 27748–27756 [DOI] [PubMed] [Google Scholar]
- 36. McBride A. A., Oliveira J. G., McPhillips M. G. (2006) Partitioning viral genomes in mitosis. Same idea, different targets. Cell Cycle 5, 1499–1502 [DOI] [PubMed] [Google Scholar]
- 37. Lee A. Y., Chiang C. M. (2009) Chromatin adaptor Brd4 modulates E2 transcription activity and protein stability. J. Biol. Chem. 284, 2778–2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McBride A. A., Romanczuk H., Howley P. M. (1991) The papillomavirus E2 regulatory proteins. J. Biol. Chem. 266, 18411–18414 [PubMed] [Google Scholar]
- 39. Webster K., Parish J., Pandya M., Stern P. L., Clarke A. R., Gaston K. (2000) The human papillomavirus (HPV) 16 E2 protein induces apoptosis in the absence of other HPV proteins and via a p53-dependent pathway. J. Biol. Chem. 275, 87–94 [DOI] [PubMed] [Google Scholar]
- 40. Bellanger S., Tan C. L., Xue Y. Z., Teissier S., Thierry F. (2011) Tumor suppressor or oncogene? A critical role of the human papillomavirus (HPV) E2 protein in cervical cancer progression. Am. J. Cancer Res. 1, 373–389 [PMC free article] [PubMed] [Google Scholar]
- 41. Pfefferle R., Marcuzzi G. P., Akgül B., Kasper H. U., Schulze F., Haase I., Wickenhauser C., Pfister H. (2008) The human papillomavirus type 8 E2 protein induces skin tumors in transgenic mice. J. Invest. Dermatol. 128, 2310–2315 [DOI] [PubMed] [Google Scholar]
- 42. Hufbauer M., Lazić D., Akgül B., Brandsma J. L., Pfister H., Weissenborn S. J. (2010) Enhanced human papillomavirus type 8 oncogene expression levels are crucial for skin tumorigenesis in transgenic mice. Virology 403, 128–136 [DOI] [PubMed] [Google Scholar]
- 43. Leykauf K., Kabsch K., Gassler N., Gissmann L., Alonso A., Schenkel J. (2008) Expression of the HPV11 E2 gene in transgenic mice does not result in alterations of the phenotypic pattern. Transgenic Res. 17, 1–8 [DOI] [PubMed] [Google Scholar]
- 44. Lin K. Y., Guarnieri F. G., Staveley-O'Carroll K. F., Levitsky H. I., August J. T., Pardoll D. M., Wu T. C. (1996) Treatment of established tumors with a novel vaccine that enhances major histocompatibility class II presentation of tumor antigen. Cancer Res. 56, 21–26 [PubMed] [Google Scholar]
- 45. Blachon S., Bellanger S., Demeret C., Thierry F. (2005) Nucleo-cytoplasmic shuttling of high risk human Papillomavirus E2 proteins induces apoptosis. J. Biol. Chem. 280, 36088–36098 [DOI] [PubMed] [Google Scholar]
- 46. Liu Q., Paroo Z. (2010) Biochemical principles of small RNA pathways. Annu. Rev. Biochem. 79, 295–319 [DOI] [PubMed] [Google Scholar]
- 47. Chen L. L., Carmichael G. G. (2010) Decoding the function of nuclear long non-coding RNAs. Curr. Opin. Cell Biol. 22, 357–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang Q., Carmichael G. G. (2004) Effects of length and location on the cellular response to double-stranded RNA. Microbiol. Mol. Biol. Rev. 68, 432–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wahid A. M., Coventry V. K., Conn G. L. (2008) Systematic deletion of the adenovirus-associated RNAI terminal stem reveals a surprisingly active RNA inhibitor of double-stranded RNA-activated protein kinase. J. Biol. Chem. 283, 17485–17493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hellwig S., Bass B. L. (2008) A starvation-induced noncoding RNA modulates expression of Dicer-regulated genes. Proc. Natl. Acad. Sci. U.S.A. 105, 12897–12902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hawley-Nelson P., Vousden K. H., Hubbert N. L., Lowy D. R., Schiller J. T. (1989) HPV16 E6 and E7 proteins cooperate to immortalize human foreskin keratinocytes. EMBO J. 8, 3905–3910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gewin L., Galloway D. A. (2001) E box-dependent activation of telomerase by human papillomavirus type 16 E6 does not require induction of c-myc. J. Virol. 75, 7198–7201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Helt A. M., Galloway D. A. (2001) Destabilization of the retinoblastoma tumor suppressor by human papillomavirus type 16 E7 is not sufficient to overcome cell cycle arrest in human keratinocytes. J. Virol. 75, 6737–6747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Liu X., Roberts J., Dakic A., Zhang Y., Schlegel R. (2008) HPV E7 contributes to the telomerase activity of immortalized and tumorigenic cells and augments E6-induced hTERT promoter function. Virology 375, 611–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Villegas J., Müller I., Arredondo J., Pinto R., Burzio L. O. (2002) A putative RNA editing from U to C in a mouse mitochondrial transcript. Nucleic Acids Res. 30, 1895–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Simpson L., Aphasizhev R., Gao G., Kang X. (2004) Mitochondrial proteins and complexes in Leishmania and Trypanosoma involved in U-insertion/deletion RNA editing. RNA 10, 159–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rogers K., Gao G., Simpson L. (2007) Uridylate-specific 3′ 5′-exoribonucleases involved in uridylate-deletion RNA editing in trypanosomatid mitochondria. J. Biol. Chem. 282, 29073–29080 [DOI] [PubMed] [Google Scholar]
- 58. Kapushoc S. T., Simpson L. (1999) In vitro uridine insertion RNA editing mediated by cis-acting guide RNAs. RNA 5, 656–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Nakahara T., Lambert P. F. (2007) Induction of promyelocytic leukemia (PML) oncogenic domains (PODs) by papillomavirus. Virology 366, 316–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lallemand-Breitenbach V., de Thé H. (2010) PML nuclear bodies. Cold Spring Harb. Perspect. Biol. 2, a000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Guccione E., Lethbridge K. J., Killick N., Leppard K. N., Banks L. (2004) HPV E6 proteins interact with specific PML isoforms and allow distinctions to be made between different POD structures. Oncogene 23, 4662–4672 [DOI] [PubMed] [Google Scholar]
- 62. Swindle C. S., Zou N., Van Tine B. A., Shaw G. M., Engler J. A., Chow L. T. (1999) Human papillomavirus DNA replication compartments in a transient DNA replication system. J. Virol. 73, 1001–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bischof O., Nacerddine K., Dejean A. (2005) Human papillomavirus oncoprotein E7 targets the promyelocytic leukemia protein and circumvents cellular senescence via the Rb and p53 tumor suppressor pathways. Mol. Cell Biol. 25, 1013–1024 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 64. Ebert M. S., Sharp P. A. (2010) MicroRNA sponges. Progress and possibilities. RNA 16, 2043–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P. P. (2011) A ceRNA hypothesis. The Rosetta Stone of a hidden RNA language? Cell 146, 353–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Cazalla D., Yario T., Steitz J. A. (2010) Down-regulation of a host microRNA by a herpesvirus saimiri noncoding RNA. Science 328, 1563–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Xue Y., Bellanger S., Zhang W., Lim D., Low J., Lunny D., Thierry F. (2010) HPV16 E2 is an immediate early marker of viral infection, preceding E7 expression in precursor structures of cervical carcinoma. Cancer Res. 70, 5316–5325' [DOI] [PubMed] [Google Scholar]
- 68. Lewis B. P., Burge C. B., Bartel D. P. (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA target. Cell 120, 15–20 [DOI] [PubMed] [Google Scholar]