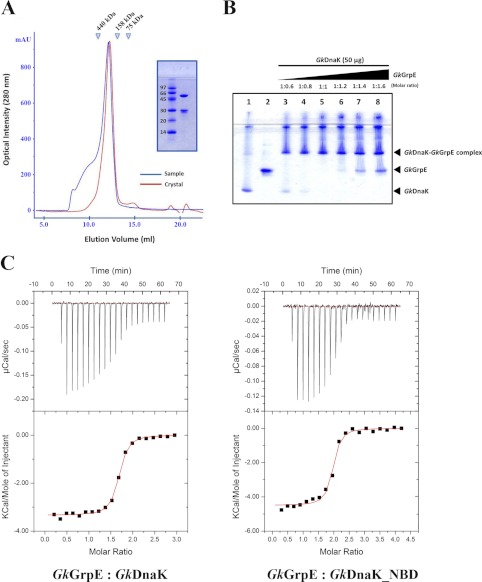
FIGURE 2.
Stoichiometry of GkDnaK with GkGrpE homodimer. A, gel filtration analysis of GkDnaK-GkGrpE complex. The elution profiles colored in blue and in red represent the complex protein solution before crystallization and the complex solution dissolved from protein crystals, respectively. The x-axis and y-axis represent the elution volume and optical intensity by UV spectrometer at 280 nm, respectively. Inset, SDS-PAGE analysis of GkDnaK-GkGrpE complex dissolved from crystals. B, interprotein interactions GkDnaK and GkGrpE performed by increasing concentrations of GkGrpE protein at different molar ratios (lanes 3-8) as indicated with control proteins (protein sample do not contain partner protein; lanes 1 and 2). The individual bands corresponding to GkDnaK and GkGrpE were observed to disappear at equimolar ratio (lane 5). A further increase in GkGrpE concentration (lanes 6-8) over GkDnaK does not result in further binding indicating that the interaction follows 2:2 stoichiometry, not 2:1 as speculated earlier in Gk species. C, upper panels, heat release/s after addition of aliquots of the GkGrpE homodimer into a calorimetry cell containing GkDnaK or GkDnaK_NBD. Lower panels, integrated binding isotherms (black circles, derived from upper panel) and experimental fits (solid red lines) to a single-site model. The best-fit molar-binding stoichiometry values are 1.59 and 1.88 for GkGrpE with GkDnaK and GkDnaK_NBD, respectively.