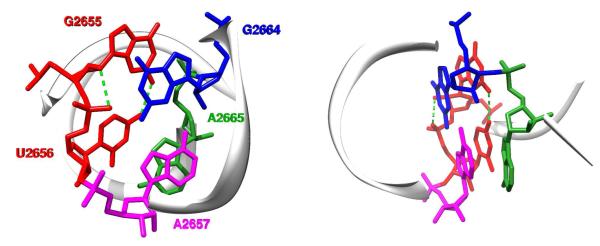
Figure 3.
Image of the sarcin/ricin domain in the ultra high-resolution (1.00 Å) structure of Escherichia coli 23S rRNA36 (PDB ID: 3dvz). The highly conserved asymmetric GpUpA/GpA miniduplex is depicted at the atomic level using a stick representation. Left, view along the duplex helix axis; right, side view. The GpU dinucleotide platform (G2655pU2656) is colored in red and its two key N2(G)…O4(U) and O2′(G)…O2P(U) H-bonds are depicted by green dashed lines. For the sake of clarity, the remainder of the domain is represented by a backbone trace and numerous other H-bonds stabilizing the GpUpA/GpA miniduplex are not depicted (see Figure 2 of Ref. 33 for details). Among them, interactions of A2665 (green) from the opposite strand with the GpU platform (red) form a characteristic in-plane nucleobase triad. The GpU…A2665 in-plane arrangement is stabilized by U…A interbase H-bonding (trans Watson-Crick/Hoogsteen pattern1,2) and G…A base-phosphate interaction (4BPh class,5 see below). Note that the G2655 base of the GpU platform is bulged out of the noncanonical RNA double helix. The remaining two bases, G2664 (blue) and A2657 (magenta), form a sheared GA base pair that stacks on the triad and completes the miniduplex motif.