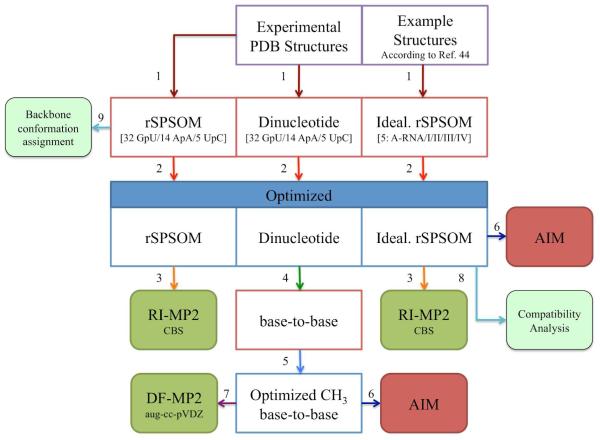
Figure 6.
A symbolic flowchart describing the complete sequence of computations. The numbered steps, denoted by arrows, are as follows: (1) extraction of model systems (numerals in parentheses in the second row of boxes denote the number of systems); (2) M06/6-31+G(d,p) and subsequent TPSS-D/LP constrained geometrical optimizations; (3) calculation of RI-MP2/CBS single-point energies; (4) removal of the sugar-phosphate backbone segment followed by attachment of terminal methyl groups to N1/N9 of pyrimidines/purines; (5) relaxation of methyl groups at the M06/6-31+G(d,p) level of theory; (6) location of H-bonds via AIM analysis; (7) evaluation of DF-MP2/aug-cc-pVDZ base…base interaction energies; (8) manual attachment of nucleobases to optimized idealized rSPSOMs and inspection of mutual compatibility to form a dinucleotide platform submotif; and (9) assignment of backbone conformational class. The experimental structures out of which idealized rSPSOMs were derived are listed in the footnote of Table 1.