Abstract
Brassinosteroids (BRs) are a group of steroidal hormones involved in plant development. Although the BR biosynthesis pathways are well characterized, the BR inactivation process, which contributes to BR homeostasis, is less understood. Here, we show that a member of the BAHD (for benzylalcohol O-acetyltransferase, anthocyanin O-hydroxycinnamoyltransferase, anthranilate N-hydroxycinnamoyl/benzoyltransferase, and deacetylvindoline 4-O-acetyltransferase) acyltransferase family may play a role in BR homeostasis in Arabidopsis (Arabidopsis thaliana). We isolated two gain-of-function mutants, brassinosteroid inactivator1-1Dominant (bia1-1D) and bia1-2D, in which a novel BAHD acyltransferase-like protein was transcriptionally activated. Both mutants exhibited dwarfism, reduced male fertility, and deetiolation in darkness, which are typical phenotypes of plants defective in BR biosynthesis. Exogenous BR treatment rescued the phenotypes of the bia1-1D mutant. Endogenous levels of BRs were reduced in the bia1-1D mutant, demonstrating that BIA1 regulates endogenous BR levels. When grown in darkness, the bia1 loss-of-function mutant showed a longer hypocotyl phenotype and was more responsive to exogenous BR treatment than the wild-type plant. BIA1 expression was predominantly observed in the root, where low levels of BRs were detected. These results indicate that the BAHD acyltransferase family member encoded by BIA1 plays a role in controlling BR levels, particularly in the root and hypocotyl in darkness. Taken together, our study provides new insights into a mechanism that maintains BR homeostasis in Arabidopsis, likely via acyl conjugation of BRs.
Plant growth and development are regulated by various environmental and endogenous biological factors. Phytohormones are important endogenous factors that control plant growth, and even small changes in their concentration and transport may have dramatic effects on plant development. Therefore, mechanisms for fine tuning the hormone concentrations in plants are critical for normal plant growth and development.
Brassinosteroids (BRs) are plant-specific steroidal hormones that are involved in many developmental processes in plants, including cell division, cell elongation, vascular differentiation, and photomorphogenesis (Clouse et al., 1996; Fujioka et al., 1997; Azpiroz et al., 1998; Choe et al., 1998; Yamamoto et al., 2007). The first identified BR, brassinolide (BL), was discovered in the pollen of Brassica napus (Grove et al., 1979). Since then, more than 70 BR compounds have been isolated from various plant species (Bajguz, 2007). During the last decade, molecular genetics and biochemical approaches have facilitated our understanding of the BR biosynthesis pathway. Loss-of-function mutants of BR-biosynthetic enzymes generally exhibit severe dwarfism, round and dark-green leaves, delayed senescence, reduced male fertility, and defective skotomorphogenesis in darkness. Mutant screening in Arabidopsis (Arabidopsis thaliana) has revealed various enzymes involved in BR biosynthesis, including the proteins encoded by the DEETIOLATED2 (Li et al., 1996), DWARF1/DIMINUTO (DWF1; Choe et al., 1999a), DWF3/CONSTITUTIVE PHOTOMORPHOGENIC DWARF (CPD; Mathur et al., 1998), DWF4 (Choe et al., 1998), DWF5 (Choe et al., 2000), DWF7/STEROL1 (Choe et al., 1999b), BR-6-oxidase1 (BR6ox1; Shimada et al., 2001), BR6ox2 (Shimada et al., 2003), and ROTUNDIFOLIA3 (ROT3; Kim et al., 2005a). Endogenous BRs regulate the expression of DWF4, CPD, BR6ox1, and ROT3 to maintain adequate BR levels, in a negative feedback loop (Goda et al., 2002; Müssig et al., 2002). BRASSINAZOLE-RESISTANT1 encodes a transcriptional repressor that inhibits the transcription of DWF4 and CPD by directly interacting with their promoter regions (He et al., 2005). DWF4 and CPD function in steroid C-22α and C-23α hydroxylations, and these two steps were recognized as the putative rate-limiting steps controlling the appropriate concentrations of endogenous BRs (Kim et al., 2006).
In addition to being controlled at the level of biosynthesis, the levels of endogenous BRs are also thought to be controlled by catabolic inactivation, including epimerization, esterification, glycosylation, hydroxylation, and sulfonation (Fujioka and Yokota, 2003). Although more than 40 BR metabolites were identified in various feeding experiments, only a few enzymes were found to be involved in the BR metabolism (Fujioka and Yokota, 2003). In Arabidopsis, the first enzyme known to be involved in the BR metabolism was isolated in a study investigating the gain-of-function phyb-4 activation-tagged suppressor1-D (bas1-D) mutant (Neff et al., 1999; Turk et al., 2003). The bas1-D mutant showed reduced hypocotyl lengths due to a decrease in bioactive BR levels. BAS1 encodes a cytochrome P450 (CYP734A1), and this hydroxylase is involved in the conversion of castasterone (CS) and BL to their C-26-hydroxylated compounds. Another cytochrome P450, CYP72C1, was identified independently by three groups through studies of chibi2 (chi2; Nakamura et al., 2005), shrinked1-D (shk1-D; Takahashi et al., 2005), and suppressor of phyb-4 7-D (sob7-D; Nakamura et al., 2005; Takahashi et al., 2005; Turk et al., 2005). CYP72C1 shares 36% amino acid identity with BAS1. On the basis of the mutant phenotypes and the results obtained from biochemical studies, CYP72C1 was predicted to function in a unique BR inactivation step that is distinct from that of BAS1 (Thornton et al., 2010).
UDP-glycosyltransferase 73C5 (UGT73C5) was identified as an enzyme involved in the glycosylation of BRs (Poppenberger et al., 2005). Plants overexpressing UGT73C5 displayed dwarfism, a typical phenotype of BR-deficient mutants, and this dwarfism was rescued by exogenous treatment with 24-epibrassinolide (24-epiBL). Biochemical liquid chromatography/mass spectrometry (LC/MS) analysis revealed that UGT73C5 converts BL into BL-23-O-glucoside. Yuan et al. (2007) isolated the bri1-5 enhanced 1-1dominant (ben1-1D) mutant and demonstrated that it contains activated dihydroflavonol 4-reductase-like protein. BEN1 was predicted to be involved in either the BR epimerization step or to modulate BR levels indirectly.
With the exception of BAS1 and CYP72C1, which are involved in the hydroxylation of BRs, and UGT73C5, which is involved in the glycosylation of BRs, enzymes involved in other BR inactivation processes remain unknown.
To identify a novel BR-metabolic pathway, we isolated and characterized dominant mutants of BRASSINOSTEROID INACTIVATOR1 (BIA1), which has an inhibitory effect on BR biosynthesis. BIA1 encodes a member of the BAHD (for benzylalcohol O-acetyltransferase, anthocyanin O-hydroxycinnamoyltransferase, anthranilate N-hydroxycinnamoyl/benzoyltransferase, and deacetylvindoline 4-O-acetyltransferase) acyltransferase family (D’Auria, 2006). Overexpression of BIA1 reduced the levels of a subset of BR compounds in the BR biosynthesis pathways and altered the expression of genes encoding BR-biosynthetic enzymes. These data suggest that the BAHD acyltransferase family member encoded by BIA1 plays a role in a novel BR inactivation process, possibly through acyl conjugation, in Arabidopsis.
RESULTS
Isolation of Two Dwarf Mutants (bia1-1D and bia1-2D) That Are Phenocopies of BR-Deficient Mutants
We generated approximately 80,000 activation-tagging lines in an effort to isolate gain-of-function mutants that showed developmental abnormalities (Ahn et al., 2007). Among them, we identified two independent gain-of-function mutants, bia1-1D and bia1-2D, that exhibited small, round, dark-green leaves, reduced stature, and delayed senescence (Fig. 1A). The BASTA (phosphinothricin) resistance trait, imparted by the bar gene in the transformation vector, cosegregated with the dwarf phenotype in these two mutants. Furthermore, DNA gel-blot analysis using the transferred (T)-DNA as a probe revealed a single band (data not shown) and each of the self-pollinated hemizygous plants generated a 3:1 segregation ratio of plants with dwarf to normal heights, suggesting that a single, semidominant locus confers the mutant phenotype. At the mature stage, the heights of both of these mutants were dramatically reduced (Fig. 1B). In addition, vascular differentiation was limited in the mutants (Fig. 1, C and D) and hypocotyl growth was reduced in both light and dark conditions (Fig. 1, E–H). The overall phenotypes displayed by the bia1-1D and bia1-2D mutants resembled those of BR-deficient mutants, such as dwf4 (Choe et al., 1998).
Figure 1.
Phenotypes of the bia1-1D and bia1-2D mutants. A, Three-week-old wild-type (WT; left), bia1-1D (center), and bia1-2D (right) plants. Scale bar = 1 cm. B, Six-week-old wild-type (WT; left), bia1-1D (center), and bia1-2D (right) plants. Insets show a magnified view of the bia1-1D and bia1-2D mutants. Scale bar = 5 mm. C, Transverse section showing vascular bundle differentiation in the wild-type (WT) plant. Scale bar = 20 μm. D, Transverse section showing vascular bundle differentiation in the bia1-1D mutant. Scale bar = 20 μm. E, Five-day-old wild-type (WT; left), bia1-1D (center), and bia1-2D (right) plants grown in white light. Scale bar = 1 mm. F, Measurement of the hypocotyl lengths of the 5-d-old plants shown in E. n ≥ 15, and data are mean ± sd values. G, Six-day-old wild-type (WT; left), bia1-1D (center), and bia1-2D (right) plants grown in darkness. Scale bar = 1 mm. H, Measurement of the hypocotyl lengths of the 6-d-old plants shown in G. n ≥ 15, and data are mean ± sd values.
BR Signaling Was Not Altered in the bia1-1D Mutant
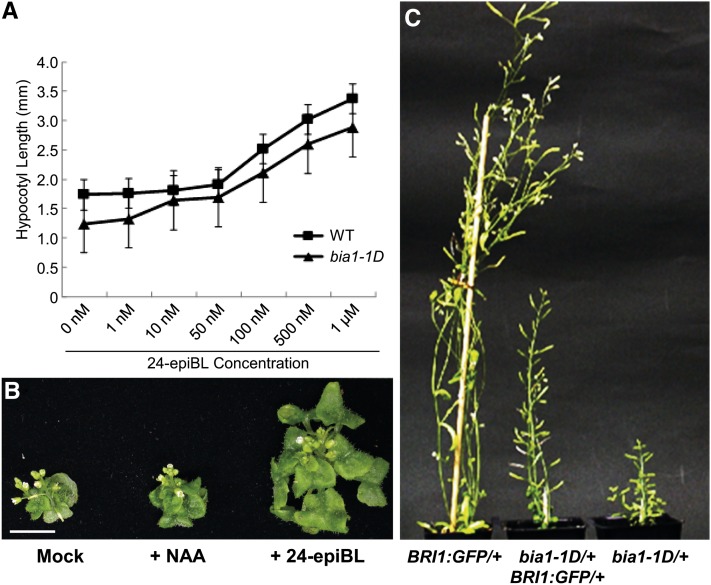
In light conditions, exogenous treatment of 24-epiBL is known to result in hypocotyl elongation (Nemhauser et al., 2003). To test whether the bia1-1D mutant was responsive to exogenous BR, we analyzed the phenotypes of wild-type and bia1-1D seedlings grown on Murashige and Skoog agar medium supplemented with various concentrations of 24-epiBL. The hypocotyl lengths of wild-type and bia1-1D plants were similar when seedlings were treated with 10 and 50 nm 24-epiBL (Fig. 2A). To determine whether the results of this seedling feeding experiment could also be applied to soil-grown mature plants, 3-week-old, soil-grown bia1-1D plants were sprayed with 1 μm of 24-epiBL every 3 d for 15 d. This treatment caused leaf expansion in the bia1-1D mutant, whereas treatment with 1 μm of synthetic auxin (l-napthaleneacetic acid) did not (Fig. 2B). Both results suggest that the dwarfism exhibited by the bia1 gain-of-function mutant was caused by a deficiency of active BRs.
Figure 2.
The shortened hypocotyl length and dwarfism of the bia1-1D mutant could be rescued by treatment with exogenous 24-epiBL or introduction of BRI1::GFP. A, Measurement of the hypocotyl length of wild-type (WT) and bia1-1D seedlings grown on plates supplemented with various concentrations of 24-epiBL. n ≥ 15, and data are mean ± sd values. B, Phenotypic rescue of the soil-grown bia1-1D mutant by periodic spraying with 24-epiBL. Scale bar = 5 mm. C, Partial rescue of dwarfism of the bia1-1D mutant by introduction of BRI1::GFP. Phenotypes of each hemizygous plant are shown for comparison.
To confirm that BR sensing was not altered in the bia1-1D mutant, we crossed the bia1-1D plant with the BRI1:GFP plant, in which the BR response was enhanced due to the presence of additional copies of the BR receptor (Wang et al., 2001). In the F1 generation, bia1-1D/+ BRI1:GFP/+ plants were of intermediate height, attaining heights that were midway between those of the bia1-1D/+ and BRI1:GFP/+ hemizygous plants. Similarly, the size of the rosette leaves was partially recovered in the bia1-1D/+ BRI1:GFP/+ plant as compared to the parental bia1-1D plant (Fig. 2C). This partial rescue suggests that the bia1-1D phenotype arises from an alteration in BR biosynthesis, and not from an alteration in BR signaling.
Up-Regulation of At4g15400 Is Responsible for the Dwarf Phenotype of the bia1-1D and bia1-2D Mutants
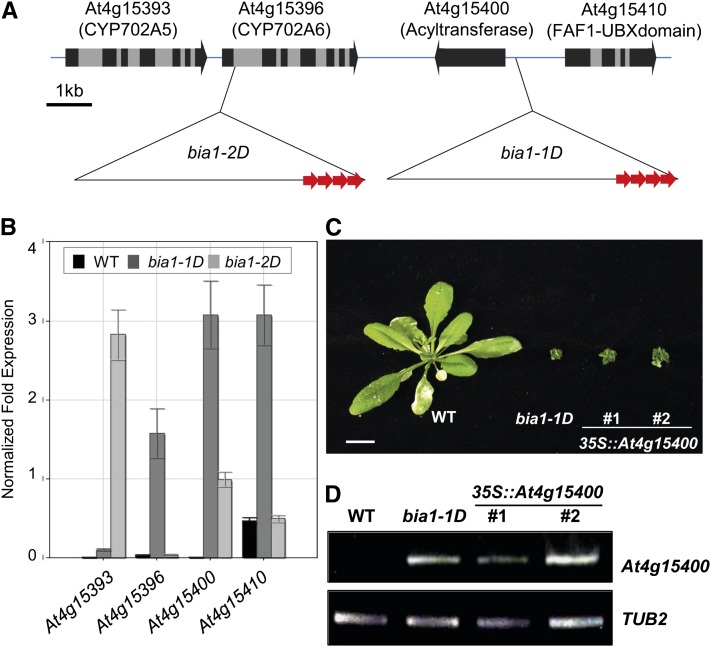
Plasmid rescue methods (Weigel et al., 2000) were employed to identify the T-DNA insertion locus in the bia1-1D and bia1-2D mutants. Sequence analysis of cloned flanking genomic DNAs revealed that the T-DNA had inserted in the upstream region of At4g15400 (approximately 600 bp from the start codon) in the bia1-1D mutant, and in the first intron of At4g15396 in the bia1-2D mutant (Fig. 3A). We then evaluated the expression of At4g15400, At4g15396, and two neighboring genes (Fig. 3A) in each of the mutants using real-time quantitative reverse transcription (qRT)-PCR. Transcript levels of At4g15393, At4g15396, At4g15400, and At4g15410 were elevated in the bia1-1D mutant, whereas those of At1g15393 and At4g15400 were up-regulated in the bia1-2D mutant (Fig. 3B).
Figure 3.
Up-regulation of At4g15400 causes dwarfism of the bia1-1D and bia1-2D mutants. A, T-DNA insertion loci in the bia1-1D and bia1-2D mutants. The three copies of the 35S enhancers on the T-DNA are marked with red arrows and gray boxes show introns. B, Real-time qRT-PCR of genes adjacent to the T-DNA insertions. The quantified transcript levels were normalized with TUB2. Error bars represent sd of three replicates. C, Recapitulated phenotype of the CaMV35S::At4g15400 transgenic plants. Scale bar = 1 cm. D, RT-PCR of BIA1 in wild-type (WT), bia1-1D, and CaMV35S::At4g15400 (number 1 and 2) plants. Twenty-two PCR cycles were performed for both At4g15400 and TUB2.
To identify the gene(s) responsible for the dwarf phenotype in these mutants, we performed a recapitulation assay. At4g15393, At4g15396, At4g15400, and At4g15410 were individually overexpressed in Arabidopsis plants under the cauliflower mosaic virus (CaMV) 35S promoter. We found that two independent transgenic plants overexpressing At4g15400 effectively phenocopied the dwarf phenotype of the bia1-1D mutant (Fig. 3C). In contrast, transgenic plants overexpressing At4g15393, At4g15396, or At4g15410 did not show any altered morphology (data not shown). We confirmed the transcriptional up-regulation of At4g15400 in the two independent CaMV35S::At4g15400 plants (Fig. 3D). These results verify that the dwarf phenotype of the bia1-1D and bia1-2D mutants is conferred by overexpression of At4g15400. Hereafter, we designate At4g15400 as BIA1.
BIA1 Encodes a Novel BAHD Acyltransferase-Like Protein
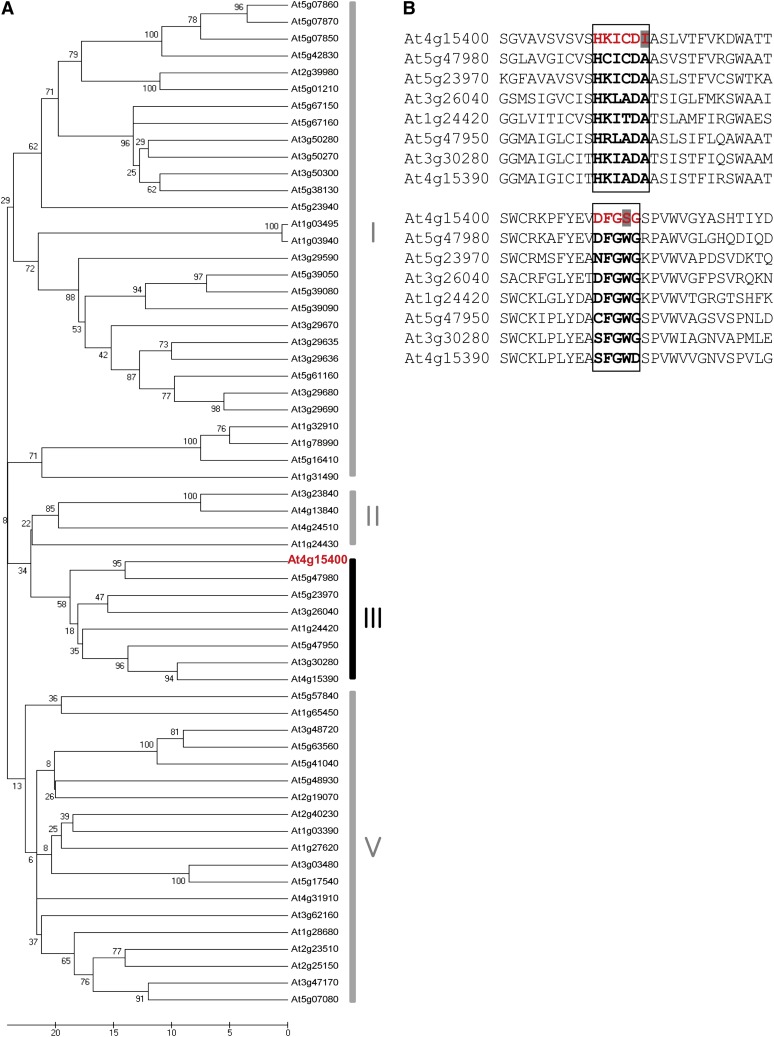
BIA1 encodes a protein consisting of 435 amino acids. Database analyses revealed that BIA1 belongs to a member of the putative BAHD family of acyltransferases, which utilize CoA thioesters and catalyze the formation of ester or amide bonds in plant metabolites (D’Auria, 2006). The Arabidopsis genome contains 61 recognized BAHD family members (Yu et al., 2009). The BAHD family can be classified into five clades through phylogenetic analysis (D’Auria, 2006), and BIA1 is a member of clade III (Fig. 4A). Some enzymes in this clade are known to utilize acetyl-CoA as the major acyl donor and are involved in the production of volatile esters in some plant species (Dudareva et al., 1998; Nam et al., 1999; Shalit et al., 2003; Beekwilder et al., 2004; El-Sharkawy et al., 2005). However, to date, none of the genes of this clade have been functionally characterized in Arabidopsis.
Figure 4.
At4g15400 encodes a BAHD acyltransferase-like protein. A, Phylogenetic tree based on the predicted amino acid sequences of At4g15400 (red) and all other BAHD acyltransferases in Arabidopsis. Clade IV does not exist in Arabidopsis. Amino acids were aligned using the Clustal W program and a phylogenetic tree was constructed using MEGA 5.05 software by scoring the number of amino acid differences (pairwise distance). Scale bar indicates the number of amino acid differences. B, Comparison of highly conserved motifs in BIA1 and other proteins in clade III of the BAHD family in Arabidopsis. Bold characters in the squares represent two conserved motifs. Amino acids of At4g15400 that differ from those of other BAHD family proteins are shaded. Red font indicates conserved motifs in BIA1.
The BAHD enzymes contain highly conserved common domains (D’Auria, 2006). The first is the HXXXDG motif (also frequently present as HXXXDA), located near the center of these proteins, and the second is the DFGWG motif, located near the C terminus of these proteins (D’Auria, 2006). BIA1 also has these two motifs; the HKICDI sequence is found in amino acids 151 to 156, with Ile instead of Ala, and the sequence DFGSG is found in amino acids 369 to 373, with Ser rather than Trp (Fig. 4B). Whether these two amino acid differences in BIA1 confer roles that are distinct from the other members of the same BAHD acyltransferase clade remains to be investigated.
Up-Regulation of BIA1 Reduced Endogenous Levels of BRs
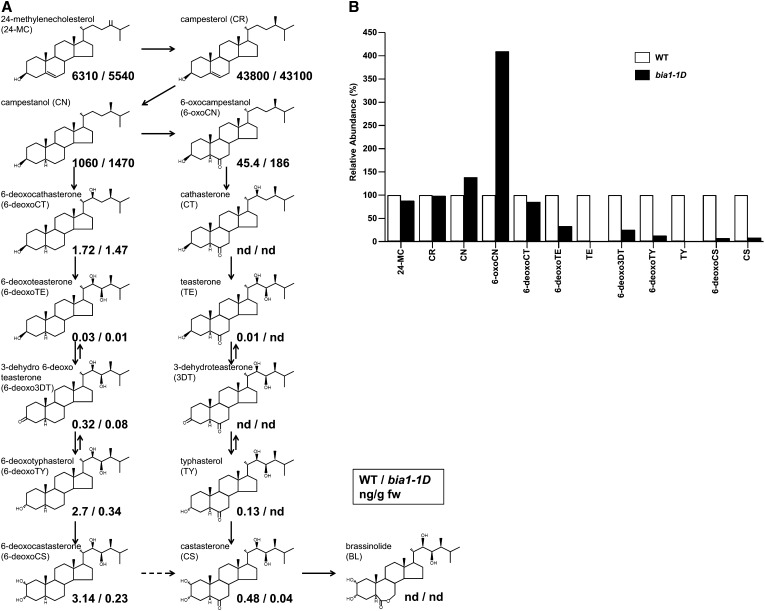
Because the bia1-1D mutant displayed phenotypes similar to those of BR-deficient mutants, we asked whether overexpression of BIA1 affected BR levels. Levels of endogenous BRs in wild-type and bia1-1D plants were analyzed by gas chromatography-mass spectroscopy (Fig. 5A). In the bia1-1D mutant, levels of 24-methylenecholesterol and campesterol were not significantly changed. However, levels of campestanol and 6-oxocampestanol increased, and those of 6-deoxoteasterone, 3-dehydro-6-deoxoteasterone, 6-deoxotyphasterol, 6-deoxocastasterone, teasterone (TE), typhasterol, and CS were reduced by various amounts (Fig. 5B). These results suggest that BIA1 can reduce endogenous levels of BRs.
Figure 5.
Quantification of endogenous BRs in wild-type and bia1-1D plants. A, BR levels measured in wild-type (WT; numbers to the left of the dash) and bia1-1D (numbers to the right of the dash) plants projected onto the proposed BR biosynthesis pathway of 24-methylenecholesterol. Numbers are in ng/g of fresh weight. B, Relative abundance of BRs in WT and bia1-1D plants. The level of each compound in the BR biosynthesis pathway in wild-type plants was arbitrarily set to 100.
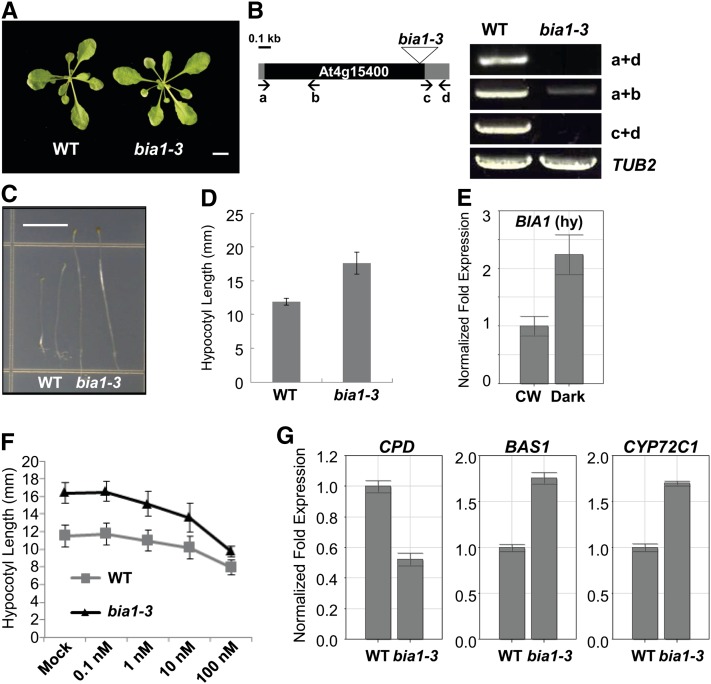
bia1-3, a Loss-of-Function Mutant of BIA1, Displayed a Longer Hypocotyl Phenotype in Dark Conditions
To analyze the phenotype of the loss-of-function mutant of BIA1, we obtained a T-DNA insertion line, bia1-3 (WiscDsLox474E11), from the Arabidopsis Biological Resource Center. When grown on soil, the bia1-3 plants did not show any obvious phenotype in the aerial parts as compared to the wild-type plants (Fig. 6A). Genotyping PCR revealed that the bia1-3 mutant has a T-DNA at the 3′ end of At4g15400. The full-length mRNA of At4g15400 was undetectable in the bia1-3 mutant. Furthermore, the 3′ end region of the At4g15400 transcript, which does not span the T-DNA, was also not detected. Although the 5′ region of At4g15400 mRNA was weakly detected in the bia1-3 plants, its levels were significantly lower than those of wild-type plants (Fig. 6B). These results indicate that the bia1-3 mutant is a near RNA-null allele of At4g15400.
Figure 6.
Isolation of bia1-3, a loss-of-function mutant of BIA1. A, Vegetative morphology of 3-week-old wild-type (WT) and bia1-3 plants. Scale bar = 1 cm. B, The T-DNA insertion site and expression of BIA1 in the bia1-3 mutant. The triangle represents the T-DNA inserted into the BIA1 coding region. The black square represents the coding region and the gray squares mean the 5′ and 3′ untranslated regions of BIA1. RT-PCR using primers (lowercase letters and arrows) that amplify different regions of BIA1 shows that the bia1-3 mutant is a near RNA-null allele. TUB2 was used as an internal control. The number of PCR cycles was 35 for BIA1 and 22 for TUB2, respectively. C, Phenotypes of wild-type (WT), bia1-1D, and bia1-3 plants grown under dark conditions. Scale bar = 5 mm. D, Measurement of the hypocotyl lengths of 5-d-old seedlings shown in C (n ≥ 15, data represent the mean ± sd values). E, Real-time qRT-PCR of BIA1 in the hypocotyls (hy) of 4-d-old wild-type seedlings grown under continuous white light (CW) or dark conditions. The fold expression was calculated relative to the expression level in continuous white light. Error bars represent sd of three replicates. F, Exogenous BR response test of hypocotyl elongation in bia1-3 and wild-type dark-grown seedlings. n ≥ 15, and data represent the mean ± sd values. G, Real-time qRT-PCR of BR-related genes in 6-d-old dark-grown wild-type (WT) and bia1-3 seedlings. The fold expression was calculated relative to the level of the WT. Error bars represent sd of three replicates.
We grew the bia1-3 seedlings vertically on Murashige and Skoog medium under continuous light or dark conditions. The loss-of-function mutant seedlings displayed a longer hypocotyl phenotype than the wild type in darkness (Fig. 6, C and D). By contrast, no difference was detected in continuous light (data not shown). We isolated RNA from the hypocotyls of 4-d-old seedlings grown under continuous light or dark conditions. We verified by real-time qRT-PCR that the BIA1 transcripts were more abundant in the hypocotyls of seedlings grown in dark conditions than in those grown under continuous light (Fig. 6E). Likewise, the hypocotyls of the bia1-3 mutant grown in darkness were more responsive to increased concentrations of BR than were those of the wild type (Fig. 6F).
In addition, to investigate whether the increased hypocotyl elongation in the dark-grown bia1-3 seedlings resulted from altered BR levels, we analyzed the transcript levels of BR-related genes. Real-time qRT-PCR showed that expression of CPD was reduced, while transcript levels of BAS1 and CYP72C1 were elevated in the bia1-3 mutant (Fig. 6G). Thus, these results indicate that BR levels were elevated in the dark-grown bia1-3 seedlings, and suggest that BIA1 may play a role in the down-regulation of BR levels in dark conditions.
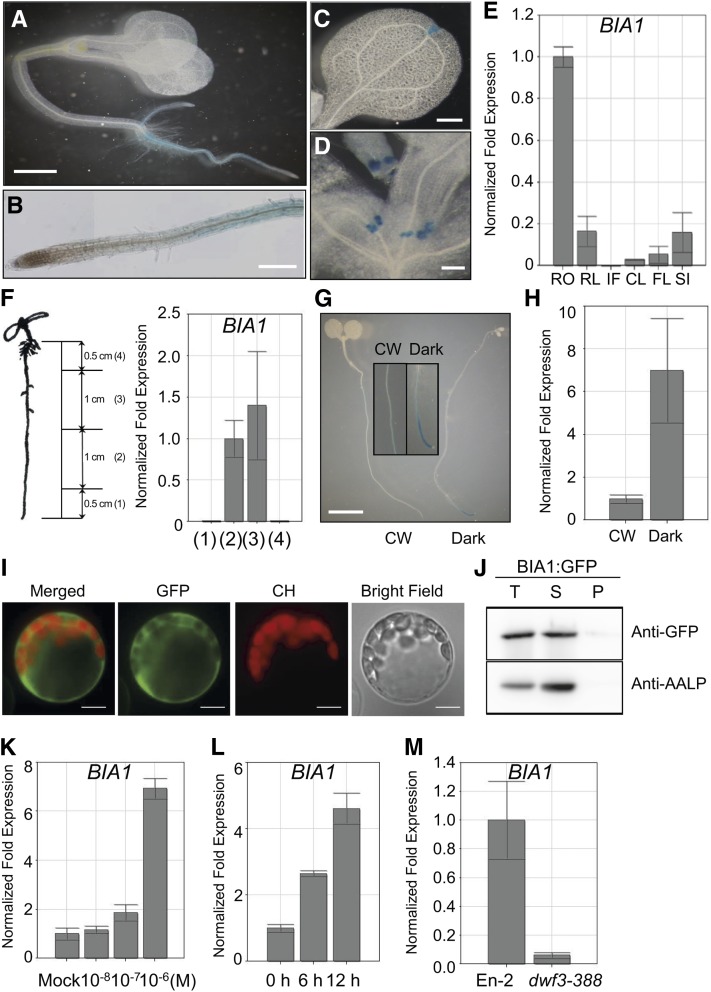
BIA1 Is Strongly Expressed in the Root Elongation Zone and Encodes a Cytosolic Acyltransferase-Like Protein
We performed histochemical GUS assays with more than 10 independent BIA1::BIA1:GUS transgenic lines to determine the spatial expression pattern of BIA1. In 5-d-old seedlings, strong GUS activity was detected in the root, especially in the elongation zone (Fig. 7, A and B). GUS activity was also detected in the hydathode of cotyledons and developing young leaves (Fig. 7, C and D). To confirm the tissue-specific expression pattern of GUS, we performed real-time qRT-PCR. Consistent with the GUS expression pattern, BIA1 expression was strongest in the root (Fig. 7E). To investigate the major zone of BIA1 expression in the root, we dissected the roots of 7-d-old plants into four sections and examined BIA1 expression in each section by real-time qRT-PCR. BIA1 was strongly expressed in the middle two sections (Fig. 7F). However, BIA1 transcripts were barely detectable in the apical section of the root (from the tip to 0.5 cm above the tip), including the root meristematic zone, and in the basal section (0.5 cm from the end of the hypocotyl), which may include the maturation zone. This result confirmed the GUS expression pattern of BIA1 (Fig. 7, A and B), indicating that BIA1 is highly expressed in the elongation zone of the root.
Figure 7.
Spatiotemporal expression and subcellular localization of BIA1. A to D, GUS staining patterns of 5- to 7-d-old BIA1::BIA1:GUS transgenic seedlings. A, BIA1::BIA1:GUS activity in a whole seedling. Scale bar = 1 mm. B, Root elongation zone and division zone. Scale bar = 100 μm. C, Cotyledon. Scale bar = 100 μm. D, Young leaf and primordia. Scale bar = 100 μm. E, Real-time qRT-PCR of BIA1 in the root (RO), rosette leaves (RL), inflorescences (IF), cauline leaves (CL), floral clusters (FL), and siliques (SI). The fold expression was calculated relative to the expression in the root. Error bars represent sd of three replicates. F, Real-time qRT-PCR of BIA1 in the dissected roots of 7-d-old wild-type seedlings. Numbers on the graph (right) correspond with those in the diagram (left). G, GUS staining patterns of 6-d-old BIA1::BIA1:GUS transgenic seedlings grown under continuous white light (CW) or dark conditions. Scale bar = 1 mm. H, Real-time qRT-PCR of BIA1 in 6-d-old seedlings grown under light or dark conditions. The fold expression was calculated relative to the level in continuous white light. Error bars represent sd of three replicates. I, Subcellular localization of BIA1 through transient expression of CaMV35S::BIA1:GFP in Arabidopsis protoplasts. CH, Chloroplast autofluorescence. Scale bar = 20 μm. J, Subcellular fractionation of BIA1:GFP isolated from Arabidopsis protoplast cells. Total protein extracts (T) were separated into soluble (S) and pellet (P) fractions. Fractions were examined by protein gel-blot analysis using anti-GFP and anti-AALP antibodies. AALP was used as a control in the fractionation procedure. K, Real-time qRT-PCR of BIA1 in 10-d-old seedlings treated with various concentrations of epi-BL for 12 h. The fold expression was calculated relative to the expression level in mock samples. Error bars represent sd of three replicates. L, Real-time qRT-PCR of BIA1 in 10-d-old wild-type seedlings treated with 1 μm 24-epiBL for 6 or 12 h. The fold expression was calculated relative to the level of samples before 24-epiBL treatment. Error bars represent sd of three replicates. M, Real-time qRT-PCR of BIA1 in wild-type (En-2) and BR-biosynthetic mutant (dwf3-388) plants. Error bars represent sd of three replicates.
To test whether BIA1 undergoes light regulation, BIA1::BIA1:GUS transgenic plants were grown in continuous light or dark conditions. We detected stronger GUS activity in the root elongation zone of the dark-grown plants than in that of the light-grown plants (Fig. 7G). We also detected transcriptional induction of BIA1 in plants grown in darkness by real-time qRT-PCR (Fig. 7H). These BIA1 expression patterns are similar to those of CYP72C1, which is involved in BR inactivation via hydroxylation in Arabidopsis (Takahashi et al., 2005).
To determine the subcellular localization of BIA1, CaMV35S::BIA1:GFP constructs were generated and transiently expressed in Arabidopsis protoplasts. Fluorescence microscopy analysis revealed that BIA1 existed in the cytosol (Fig. 7I). Immunoblotting of the protein fraction of protoplast cell extracts showed that BIA1 was present in the soluble fraction along with aleurain-like protein (AALP; Fig. 7J), which is known to be a soluble protein (Sohn et al., 2003). The cytosolic localization of BIA1 is consistent with those of 3-β-BR dehydrogenase (Stündl and Schneider, 2001) and dihydroflavonol 4-reductase-like protein (BEN1; Yuan et al., 2007), which are involved in BR metabolism, further suggesting that BIA1 functions in the cytosol.
BIA1 Is Induced by BR
Overexpression of BIA1 led to a deficiency of endogenous BRs, as observed in bas1-D, shk1-D/chi2/sob7-D, ben1-D, and UGT73C5-overexpressing plants (Neff et al., 1999; Poppenberger et al., 2005; Takahashi et al., 2005; Yuan et al., 2007). BAS1 and CYP72C1 (SHK1/CHI2/SOB7/CYP72C1), which encode known BR inactivation enzymes, are regulated by BRs to maintain BR homeostasis (Müssig et al., 2002; Jung et al., 2010). To investigate whether BIA1 is regulated by BR, similarly to other BR-inactivating genes, such as BAS1 and CYP72C1, we performed real-time qRT-PCR using 10-d-old wild-type seedlings grown in various concentrations of 24-epiBL. One micromolar 24-epiBL led to approximately a 7-fold increase in BIA1 transcript levels (Fig. 7K). Similarly, 1 μm 24-epiBL treatment for 12 h caused about a 5-fold induction of BIA1 transcripts (Fig. 7L). To further investigate whether endogenous BR levels affect BIA1 expression, we examined levels of BIA1 transcript in the dwf3-388 mutant (Kwon and Choe, 2005), which is known to have low levels of endogenous BRs. In the dwf3-388 mutant, BIA1 transcripts were down-regulated as compared to those of wild-type plants of ecotype En-2 (Fig. 7M). These results indicate that BIA1, like other BR-inactivating enzymes, may play a role in BR homeostasis.
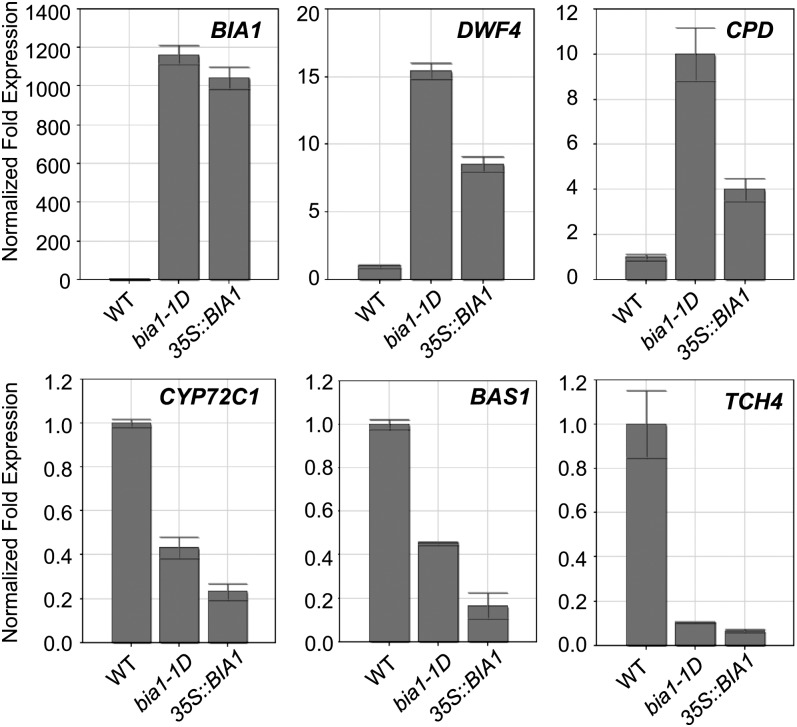
Overexpression of BIA1 Altered Expression Patterns of BR-Related Genes
DWF4 and CPD encode P450 enzymes that are involved in BR biosynthesis and are up-regulated in the BR-deficient mutants through a feedback regulation mechanism (Bancoş et al., 2002; Tanaka et al., 2005). BAS1 and CYP72C1, which inactivate bioactive BRs, were reported to be diminished in several BR-deficient mutants (Tanaka et al., 2005). TCH4, which encodes a xyloglucan endotransglucosylase/hydrolase, a cell wall modification enzyme, is repressed in BR-deficient mutants, such as det2-1 (Iliev et al., 2002). Thus, it is possible that if BIA1 metabolizes or modifies bioactive BRs into their inactive forms, the expression of DWF4 and CPD would be up-regulated and that of BAS1, CYP72C1, and TCH4 would be down-regulated in the BIA1-overexpressing plants. To test this hypothesis, we performed real-time qRT-PCR to examine the transcript levels of these genes in the bia1-1D and CaMV35S::BIA1 plants. As expected, levels of DWF4 and CPD transcripts were elevated, and those of BAS1, CYP72C1, and TCH4 were reduced in 3-week-old bia1-1D and CaMV35S::BIA1 plants (Fig. 8). These results are consistent with the observation that overexpression of BIA1 caused a decrease in endogenous levels of BRs and suggest that the BR-biosynthetic genes are transcriptionally activated to produce more BRs in the BIA1-overexpressing plants.
Figure 8.
Real-time qRT-PCR of BR-responsive genes in wild-type, bia1-1D, and CaMV35S::BIA1 plants. Real-time qRT-PCR of BIA1, DWF4, CPD, BAS1, CYP72C1, and TCH4 was carried out with RNA isolated from the aerial parts of 3-week-old wild-type (WT), bia1-1D, and CaMV35S::BIA1 plants. The fold expression was normalized relative to each gene in wild-type plants. Error bars represent sd of three replicate samples.
DISCUSSION
In this study, we characterized two activation-tagging mutants of Arabidopsis, designated as bia1-1D and bia1-2D, which exhibited dwarf phenotypes. Up-regulation of BIA1, a gene encoding a BAHD acyltransferase-like protein, reduced endogenous levels of active BRs and resulted in dwarf phenotypes for both mutants. These results suggest that BIA1 may be involved in controlling the levels of active BRs, most likely through acylation, in Arabidopsis.
BIA1 Is Involved in BR Metabolism in Arabidopsis
We have provided evidence that BIA1 is involved in the BR metabolic process in Arabidopsis. First, two activation-tagging mutants, bia1-1D and bia1-2D, and transgenic plants overexpressing BIA1 displayed dwarf phenotypes similar to other BR-deficient mutants, such as dwf4 (Choe et al., 1998), as shown in Figure 1. In addition, when the bia1-1D mutant was crossed with a transgenic plant overexpressing the BR receptor gene, BRI1, an intermediate phenotype was observed in the F1 generation (Fig. 2C). These data suggest that the two activation-tagging mutants did not exhibit defects in BR signal transduction, but instead had deficiencies in active BRs. Second, analysis of the BR profile revealed that major BRs, such as TE, typhasterol, and CS, were drastically reduced in the bia1 gain-of-function mutant (Fig. 5). Accordingly, the shortened hypocotyl and dwarf phenotypes of the bia1-1D mutant were partially recovered by exogenous treatment with 24-epiBL (Fig. 2B). Third, the expression of genes that are responsible for the synthesis of BRs was altered. In BR-deficient mutants, DWF4 and CPD, which encode key enzymes in BR biosynthesis, are up-regulated, whereas BAS1, which is involved in the BR metabolic pathway, is down-regulated through a negative feedback mechanism (Choe et al., 2001). In the bia1-1D mutant and the transgenic plant overexpressing BIA1, levels of DWF4 and CPD were elevated, while those of BAS1, CYP72C1, and TCH4 were reduced (Fig. 8). Fourth, the bia1 loss-of-function mutant was more responsive to exogenous BR treatment than was the wild-type plant when grown in darkness. Finally, BIA1 was mainly expressed in the elongation zone of the root (Fig. 7, A, B, and F). The root is known to be actively involved in BR biosynthesis, as previously shown in studies that analyzed the expression of BR metabolic genes and the biochemical properties of BR intermediates (Shimada et al., 2003). Analogous tissue-specific expression patterns were detected for other BR-inactivating enzymes, such as BAS1 (Neff et al., 1999), CYP72C1 (Takahashi et al., 2005), and BEN1 (Yuan et al., 2007). Consistent with the proposed function of BIA1 in the inactivation of active BRs in the root, exogenous BR treatment induced the expression of BIA1 transcripts (Fig. 7, K and L). These results suggest that BIA1 plays a role in the metabolism of endogenous BRs.
BIA1 Is a BAHD Acyltransferase-Like Protein Involved in the Inactivation of BRs
Through feeding experiments with exogenously applied BRs and purification of natural BR metabolites, more than 40 BR metabolites have been identified from various plants (Bajguz, 2007). These metabolites can be processed by modifications of the steroidal skeletons or side chains. The metabolic processes that inactivate bioactive BRs include dehydrogenation at C-3 and C-23 (Hai et al., 1996; Watanabe et al., 2000), epimerization at C-2, C-3, and C-24 (Suzuki et al., 1993; Nishikawa et al., 1995), hydroxylation at C-20, C-25, and C-26 (Voigt et al., 1993; Kolbe et al., 1996; Yokota et al., 1996), side chain cleavage at C-20/22 (Kolbe et al., 1996), and sulfonation at C-22 (Rouleau et al., 1999). In addition, active BRs are inactivated through conjugation by esterification at C-3 (Kolbe et al., 1995; Asakawa et al., 1996) and glycosylation at C-2, C-3, C-23, C-25, and C-26 (Bajguz, 2007).
During the last decade, the activation-tagging approach has been used as an effective strategy to identify and isolate BR-inactivating enzymes. Three enzymes (BAS1, CYP72C1, and BEN1) involved in BR inactivation were identified by the activation-tagging strategy. BAS1 was discovered as an enzyme that converts CS and BL into C-26 hydroxylated compounds (Neff et al., 1999). CYP72C1 was predicted to be involved in the hydroxylation of BRs (Thornton et al., 2010), and BEN1 most likely inactivates BRs through an unknown metabolic process (Yuan et al., 2007). Using a similar strategy, glucosyl conjugation at 23-OH of BL was identified in a UGT73C5-overexpressing transgenic line (Poppenberger et al., 2005). In addition to these four enzymes in Arabidopsis, a steroid sulfotransferase, BNST3, which is involved in the BR inactivation process through sulfonation at 22-OH of 24-epicathasterone in vitro, was isolated from B. napus (Rouleau et al., 1999). An Arabidopsis ortholog of BNST3, AtST1, was also characterized using a similar method (Marsolais et al., 2007). Although a few enzymes involved in hydroxylation, sulfonation, and glycosylation have been defined using those approaches, the enzymes involved in the production of many other BR metabolites remain to be identified.
BIA1 encodes a member of the BAHD acyltransferase family. BAHD acyltransferases are known to synthesize and modify various metabolites, such as shikimate-phenylpropanoid-derived phytoalexins, alkaloids, terpenoids, and polyamines (Yang et al., 1997; Walker et al., 2002; Luo et al., 2009). Given that BRs are terpenes, specifically, triterpenoids consisting of 30 carbons, BIA1 may play a role in adding an acyl moiety to BRs through acyltransferase activity. Among the BR metabolites, esterified BRs require conjugation with fatty acids by acyltransferases, because esterification is typical of the acylation process. The natural product of an esterified form of BRs, TE-3β-myristate, was isolated from Lilium longiflorum (Asakawa et al., 1994; Soeno et al., 2000). TE-3β-laurate was also detected in L. longiflorum (Asakawa et al., 1996). The esterified metabolites also include conjugates of 3,24-diepiCS and 3,24-diepiBL with acyl components, such as laurate, myristate, and palmitate in Ornithopus sativus (Kolbe et al., 1995). However, considering that esterification of CS and BL with laurate, myristate, and palmitate requires β-epimerization, TE could be a candidate for esterification by BIA1. The bia1-1D mutant showed the most severe phenotype among the gain-of-function mutants inactivating BRs that had been previously isolated (Neff et al., 1999; Poppenberger et al., 2005; Takahashi et al., 2005; Yuan et al., 2007). This coincides with our observation that BRs derived from TE or 6-deoxoteasterone were diminished in the bia1-1D mutant (Fig. 5). A deficiency in most BRs could cause the severe dwarfism observed in the bia1 gain-of-function mutants. This possibility should be further analyzed by additional feeding experiments with TE.
BIA1 Is Involved in Maintaining BR Homeostasis in the Root and Hypocotyl in the Dark
The root, where BIA1 is mainly expressed, is an active site of BR biosynthesis and catabolism. The promoter activity of CYP85A2, a gene that encodes a bifunctional enzyme with BR C-6 oxidase and BL synthase activities (Kim et al., 2005c) and that catalyzes C-6 oxidation, a rate-limiting step in BR biosynthesis, was found to be high in the root (Castle et al., 2005). Transcripts of ROT3/CYP90C1, which also encodes a protein that is involved in BR biosynthesis, are more abundant in the root than in the shoot (Bancoş et al., 2002). However, biochemical BR profiling analyses revealed that, although high levels of BR intermediates were detected in the root, only low or undetectable amounts of bioactive BRs were found in the root (Bancoş et al., 2002; Shimada et al., 2003). Because high levels of BRs inhibit elongation even at subnanomolar concentrations (Clouse et al., 1996; Clouse and Sasse, 1998), the root elongation zone has to contain appropriate levels of bioactive BRs. Therefore, BRs must be maintained at the optimal concentration to support normal growth of the root, and this homeostasis is tightly regulated by the antagonistic processes of BR biosynthesis and inactivation. BIA1 tissue expression profiles suggest that BIA1 is correlated with BR metabolism in the root elongation zone (Fig. 7, F and G).
Interestingly, we established that BIA1 is tightly coexpressed with CYP72C1, a putative gene found to encode a BR-inactivating enzyme through computational studies (Obayashi et al., 2007; http://atted.jp). Consistent with the Web-based study, BIA1 exhibited transcriptional regulation patterns similar to CYP72C1 (Nakamura et al., 2005; Takahashi et al., 2005; Turk et al., 2005). BIA1 and CYP72C1 were expressed in the elongation zone of the root (Fig. 7, F and G; Nakamura et al., 2005; Takahashi et al., 2005; Turk et al., 2005). In addition, the expression patterns of BIA1 and CYP72C1 were similar under light conditions, and both genes were induced in the dark (Figs. 6E and 7H; Nakamura et al., 2005; Takahashi et al., 2005; Turk et al., 2005). The dark-grown seedlings of the bia1 loss-of-function mutant exhibited an elongated hypocotyl (Fig. 6, C and D). These phenotypes were identical with those of the cyp72c1 loss-of-function mutant (Takahashi et al., 2005). Although GUS activity driven by the BIA1 promoter was not detectable in the hypocotyl, real-time qRT-PCR showed that BIA1 mRNAs increased in the hypocotyl when plants were grown in darkness (Fig. 6E). As shown in Figure 7, K and L, BIA1 was induced by BR, as was also previously observed for CYP72C1 (Jung et al., 2010). The dark-grown seedlings of the bia1 loss-of-function mutant were more sensitive to treatment with exogenous BR than were those of the wild type (Fig. 6F). This property of bia1-3 is opposite to that of the bas1 loss-of-function mutant that is sensitive to BR only in light (Turk et al., 2003). Therefore, BIA1 may encode a type of BR-inactivating enzymes that is under BR-dependent feedback regulation to maintain appropriate BR concentrations in Arabidopsis, particularly for the modulation of skotomorphogenesis. Another interesting finding was that CYP72C1 was also down-regulated in the bia1-1D mutant and in the BIA1-overexpressing transgenic line (Fig. 8). These results suggest that BIA1 and CYP72C1 may play cooperative roles in BR inactivation, maintaining the appropriate amounts of endogenous BRs within the plant, and this possibility needs to be further investigated.
In summary, we have discovered a novel BAHD acyltransferase-like protein that may be involved in the regulation of endogenous BR concentrations. Although BR metabolites isolated in plants include some acyl-conjugated forms of BRs (Fujioka and Yokota, 2003), the enzyme that transfers the acyl group to BRs has hitherto not been identified. Hence, BIA1 may have a novel role in the BR metabolic process by catalyzing the acyl conjugation of BRs in Arabidopsis.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 was used throughout this study, except for the dwf3-388 mutant (En-2 ecotype; Kwon and Choe, 2005). Plants were grown in soil mixed with vermiculite in small pots under long-day conditions (16 h of light at 24°C/8 h of dark at 22°C). The light intensity was 80 μmol m−2 s−1 for all experiments involving seedling growth, unless stated otherwise. Arabidopsis plants were transformed by the floral-dip method (Clough and Bent, 1998). Activation-tagging plants were selected for BASTA (phosphinothricin) resistance. The T-DNA-tagged bia1-3 allele was obtained from the Arabidopsis Biological Resource Center stock center (http://www.arabidopsis.org).
Identification of the T-DNA Locus in the bia1-1D and bia1-2D Mutants
To identify the T-DNA insertion locus in the mutants, the plasmid rescue method was employed (Weigel et al., 2000). Genomic DNA was extracted using the DNeasy plant mini kit (Qiagen). One microgram of genomic DNA was digested with BamHI for 16 h to rescue the left border of the T-DNA. Digested genomic DNA was ligated by T4 DNA ligase (Fermentas) overnight at 4°C. The ligated genomic DNA was transformed into Escherichia coli cells, and transformed colonies were selected on Luria-Bertani medium supplemented with ampicillin. Plasmids were extracted from each colony and sequenced to identify the region adjacent to the T-DNA using the primer JW01 (Supplemental Table S1). The identified T-DNA loci were confirmed by PCR genotyping, using the primers LB3 and bia1fw for the bia1-1D mutant, or bia2fw for the bia1-2D mutant (Supplemental Table S1).
Tissue Sections of Arabidopsis Internodes
Internodes were isolated from plants that were a little over 6 weeks old and were immediately immersed in formaldehyde-acetic-acid fixative solution (3.7% formaldehyde, 5% acetic acid, and 50% ethanol). The mixture of tissue and fixative solution was placed in a vacuum container (>20 atm) for 15 min. The fixed tissues were dehydrated in a graded series of ethanol and xylene over an interval of 30 min. The dehydrated tissues were embedded in paraplasts and sliced into 8-μm sections using a microtome (Leica Instruments GmbH).
Phylogenetic Tree Construction
Arabidopsis BAHD family proteins were aligned using the Clustal W program for multiple alignment. This alignment was applied to MEGA5.05 software for scoring the number of amino acid differences and for generating 1,000 bootstrap replicates. Through this analysis, a phylogenetic tree was constructed by the neighbor-joining method.
Generation of CaMV35S::BIA1, BIA1::BIA1:GUS, and CaMV35S::BIA1:GFP Plants
To generate CaMV35S::BIA1 plants, BIA1 genomic DNA was PCR amplified using primers 15400gwfw and 15400gwrv (Supplemental Table S1). This fragment was reamplified using the primer pair attB1 and attB2 (Supplemental Table S1). T1 transgenic plants were selected on Murashige and Skoog medium containing kanamycin sulfate. To generate the BIA1::BIA1:GUS construct, a region containing the 1.5-kb BIA1 promoter and full-length BIA1 cDNA was amplified using the primer pair 15400pfw and 15400crv (Supplemental Table S1). This fragment was reamplified using the adapting primer pair attB1 and attB2. The resulting amplicons for CaMV35S::BIA1 and BIA1::BIA1:GUS were cloned into the binary vector pK2GW7 containing the CaMV 35S promoter and the binary vector pKGWFS7, respectively, using Gateway BP and LR reactions (Invitrogen). For the CaMV35S::BIA1:GFP plasmid, the full-length coding region of BIA1 was amplified using primers 15400-GFPfw and 15400-GFPrv and was then subcloned into XbaI and BamHI restriction sites in the p326-GFP vector (Lee et al., 2009) for N-terminal fusion with GFP.
Hypocotyl Measurements
After surface sterilization, seeds were sown on Murashige and Skoog medium plates supplemented with 1.5% plant agarose (DUCHEFA) and with various concentrations of 24-epiBL (Sigma-Aldrich) for BR feeding experiments. Plates were cold treated at 4°C for 3 d, exposed to white light for 6 h, and then transferred into light or dark conditions at 24°C and grown vertically. After 6 d, hypocotyl lengths were measured using a LAS-3000 CCD camera and MultiGauge version 3.0 software (Fujifilm).
Real-Time qRT-PCR
Total RNAs for real-time qRT-PCR were extracted from 3-week-old plants using the RNeasy plant mini kit with on-column DNase treatment (Qiagen). Moloney murine leukemia virus reverse transcriptase (Fermentas) was used to synthesize first-strand cDNA from 2 μg of total RNA using oligo(dT). Real-time qRT-PCR was performed with the Bio-Rad iQ5 system, using the SYBR green master mix (Bio-Rad), in accordance with the manufacturer’s instructions. Expression of each sample was normalized with tubulin-2 (TUB2) expression, and normalized fold expressions were determined using iQ5 optical system software version 1.0 (Bio-Rad). All of the real-time qRT-PCRs were repeated at least three times.
At4g15400 was amplified using primers 15400-qfw and 15400-qrv; DWF4 was amplified using primers DWF4-qfw and DWF4-qrv; CPD was amplified using primers CPD-qfw and CPD-qrv; BAS1 was amplified using primers BAS1-qfw and BAS1-qrv; CYP72C1 was amplified using primers CYP72C1-qfw and CYP72C1-qrv; and TCH4 was amplified using primers TCH4-qfw and TCH4-qrv (Supplemental Table S1). All primer sets were designed to involve intron-spanning regions, except for the primers for At4g15400.
BR Profile Analysis
For BR measurements, the aerial parts of 6-week-old plants grown under long-day conditions (16 h of light at 24°C/8 h of dark at 22°C) were harvested. One hundred and twenty grams of wild type or 45 g of the bia1-1D plants were removed and then lyophilized. Lyophilized tissues were extracted twice with 500 mL methanol. Next, deuterium-labeled internal standards were added. BRs were quantified by gas chromatography-mass spectroscopy, as described previously (Fujioka et al., 2002).
Histochemical GUS-Staining Analysis
Ten independent BIA1::BIA1:GUS transgenic plants (T1 generation) were selected, and expression of GUS was analyzed in these plants. Seedlings and various organs were sampled and vacuum infiltrated in 5-bromo-4-chloro-3-indolyl-β-glucuronic acid staining solution containing 50 mm sodium phosphate buffer (pH 7.0), 2 mm each of potassium ferricyanide and ferrocyanide, 2 mm 5-bromo-4-chloro-3-indolyl-β-glucuronic acid, 10 mm EDTA, and 0.1% (v/v) Triton X-100. After incubation for 12 h at 37°C, tissues were destained several times with 70% ethanol washes.
Subcellular Localization of BIA1
The CaMV35S::BIA1:GFP plasmid was introduced into Arabidopsis protoplasts prepared from leaf mesophyll by polyethylene-glycol-mediated transformation. After incubation for 24 h at 22°C, images were taken using a cooled CCD camera coupled to a Zeiss Axioplan2 fluorescence microscope (Carl Zeiss, Inc.). The XF116 filter set (exciter, 474AF20; dichroic, 500DRLP; emitter, 510AF23) was used. To isolate cell extracts, protoplasts incubated for 24 h were treated with the freeze-and-thaw incubation method (Kim et al., 2005b) and then centrifuged at 10,000g at 4°C for 5 min. After removing the debris, fractionation into soluble and membrane fractions was performed by ultracentrifugation at 100,000g for 30 min. Fractionated protein was detected by protein gel-blot analysis using monoclonal anti-GFP antibodies (Clontech) for BIA1:GFP and polyclonal anti-AALP antibodies for the control of pellet fraction.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number NM_117628.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Primer sequences used in this study.
Supplementary Material
Acknowledgments
We thank Prof. Hyeonsook Cheong (Chosun University, Gwangju) and Prof. Sunghwa Choe (Seoul National University, Seoul) for providing BRI1:GFP and dwf3-388 seeds, respectively, and Prof. Inhwan Hwang (Pohang University of Science and Technology, Pohang) for providing the AALP antibody. We are also thankful to Dr. Suguru Takatsuto (Joetsu University of Education) for supplying deuterium-labeled internal standards and Prof. Yeonhee Choi (Seoul National University, Seoul) for critical reading of the manuscript.
Glossary
- BRs
brassinosteroids
- BL
brassinolide
- CS
castasterone
- 24-epiBL
24-epibrassinolide
- qRT
quantitative reverse transcription
- CaMV
cauliflower mosaic virus
- CaMV
cauliflower mosaic virus
- TE
teasterone
- AALP
aleurain-like protein
- T-DNA
to be defined
References
- Ahn JH, Kim J, Yoo SJ, Yoo SY, Roh H, Choi JH, Choi MS, Chung KS, Han EJ, Hong SM, et al. (2007) Isolation of 151 mutants that have developmental defects from T-DNA tagging. Plant Cell Physiol 48: 169–178 [DOI] [PubMed] [Google Scholar]
- Asakawa S, Abe H, Kyokawa Y, Nakamura S, Natsume M. (1994) Teasterone 3-myristate: a new type of brassinosteroid derivative in Lilium longiflorum anthers. Biosci Biotechnol Biochem 58: 219–220 [DOI] [PubMed] [Google Scholar]
- Asakawa S, Abe H, Nishikawa N, Natsume M, Koshioka M. (1996) Purification and identification of new acyl-conjugated teasterones in lily pollen. Biosci Biotechnol Biochem 60: 1416–1420 [Google Scholar]
- Azpiroz R, Wu Y, LoCascio JC, Feldmann KA. (1998) An Arabidopsis brassinosteroid-dependent mutant is blocked in cell elongation. Plant Cell 10: 219–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajguz A. (2007) Metabolism of brassinosteroids in plants. Plant Physiol Biochem 45: 95–107 [DOI] [PubMed] [Google Scholar]
- Bancoş S, Nomura T, Sato T, Molnár G, Bishop GJ, Koncz C, Yokota T, Nagy F, Szekeres M. (2002) Regulation of transcript levels of the Arabidopsis cytochrome p450 genes involved in brassinosteroid biosynthesis. Plant Physiol 130: 504–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beekwilder J, Alvarez-Huerta M, Neef E, Verstappen FW, Bouwmeester HJ, Aharoni A. (2004) Functional characterization of enzymes forming volatile esters from strawberry and banana. Plant Physiol 135: 1865–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castle J, Szekeres M, Jenkins G, Bishop GJ. (2005) Unique and overlapping expression patterns of Arabidopsis CYP85 genes involved in brassinosteroid C-6 oxidation. Plant Mol Biol 57: 129–140 [DOI] [PubMed] [Google Scholar]
- Choe S, Dilkes BP, Fujioka S, Takatsuto S, Sakurai A, Feldmann KA. (1998) The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22alpha-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell 10: 231–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Dilkes BP, Gregory BD, Ross AS, Yuan H, Noguchi T, Fujioka S, Takatsuto S, Tanaka A, Yoshida S, et al. (1999a) The Arabidopsis dwarf1 mutant is defective in the conversion of 24-methylenecholesterol to campesterol in brassinosteroid biosynthesis. Plant Physiol 119: 897–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Fujioka S, Noguchi T, Takatsuto S, Yoshida S, Feldmann KA. (2001) Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. Plant J 26: 573–582 [DOI] [PubMed] [Google Scholar]
- Choe S, Noguchi T, Fujioka S, Takatsuto S, Tissier CP, Gregory BD, Ross AS, Tanaka A, Yoshida S, Tax FE, et al. (1999b) The Arabidopsis dwf7/ste1 mutant is defective in the delta7 sterol C-5 desaturation step leading to brassinosteroid biosynthesis. Plant Cell 11: 207–221 [PMC free article] [PubMed] [Google Scholar]
- Choe S, Tanaka A, Noguchi T, Fujioka S, Takatsuto S, Ross AS, Tax FE, Yoshida S, Feldmann KA. (2000) Lesions in the sterol delta reductase gene of Arabidopsis cause dwarfism due to a block in brassinosteroid biosynthesis. Plant J 21: 431–443 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Clouse SD, Langford M, McMorris TC. (1996) A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol 111: 671–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD, Sasse JM. (1998) BRASSINOSTEROIDS: essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol 49: 427–451 [DOI] [PubMed] [Google Scholar]
- D’Auria JC. (2006) Acyltransferases in plants: a good time to be BAHD. Curr Opin Plant Biol 9: 331–340 [DOI] [PubMed] [Google Scholar]
- Dudareva N, D’Auria JC, Nam KH, Raguso RA, Pichersky E. (1998) Acetyl-CoA:benzylalcohol acetyltransferase—an enzyme involved in floral scent production in Clarkia breweri. Plant J 14: 297–304 [DOI] [PubMed] [Google Scholar]
- El-Sharkawy I, Manríquez D, Flores FB, Regad F, Bouzayen M, Latché A, Pech JC. (2005) Functional characterization of a melon alcohol acyl-transferase gene family involved in the biosynthesis of ester volatiles: identification of the crucial role of a threonine residue for enzyme activity. Plant Mol Biol 59: 345–362 [DOI] [PubMed] [Google Scholar]
- Fujioka S, Li J, Choi YH, Seto H, Takatsuto S, Noguchi T, Watanabe T, Kuriyama H, Yokota T, Chory J, et al. (1997) The Arabidopsis deetiolated2 mutant is blocked early in brassinosteroid biosynthesis. Plant Cell 9: 1951–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka S, Takatsuto S, Yoshida S. (2002) An early C-22 oxidation branch in the brassinosteroid biosynthetic pathway. Plant Physiol 130: 930–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujioka S, Yokota T. (2003) Biosynthesis and metabolism of brassinosteroids. Annu Rev Plant Biol 54: 137–164 [DOI] [PubMed] [Google Scholar]
- Goda H, Shimada Y, Asami T, Fujioka S, Yoshida S. (2002) Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol 130: 1319–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove MD, Spencer GF, Rohwedder WK, Mandava N, Worley JF, Warthen JD, Steffens GL, Flippen-Anderson JL, Cook JC. (1979) Brassinolide, a plant growth-promoting steroid isolated from Brassica napus pollen. Nature 281: 216–217 [Google Scholar]
- Hai T, Schneider B, Porzel A, Adam G. (1996) Metabolism of 24-epi-castasterone in cell suspension cultures of Lycopersicon esculentum. Phytochemistry 41: 197–201 [Google Scholar]
- He JX, Gendron JM, Sun Y, Gampala SS, Gendron N, Sun CQ, Wang ZY. (2005) BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307: 1634–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev EA, Xu W, Polisensky DH, Oh MH, Torisky RS, Clouse SD, Braam J. (2002) Transcriptional and posttranscriptional regulation of Arabidopsis TCH4 expression by diverse stimuli: roles of cis regions and brassinosteroids. Plant Physiol 130: 770–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JH, Lee M, Park CM. (2010) A transcriptional feedback loop modulating signaling crosstalks between auxin and brassinosteroid in Arabidopsis. Mol Cells 29: 449–456 [DOI] [PubMed] [Google Scholar]
- Kim GT, Fujioka S, Kozuka T, Tax FE, Takatsuto S, Yoshida S, Tsukaya H. (2005a) CYP90C1 and CYP90D1 are involved in different steps in the brassinosteroid biosynthesis pathway in Arabidopsis thaliana. Plant J 41: 710–721 [DOI] [PubMed] [Google Scholar]
- Kim H, Park M, Kim SJ, Hwang I. (2005b) Actin filaments play a critical role in vacuolar trafficking at the Golgi complex in plant cells. Plant Cell 17: 888–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HB, Kwon M, Ryu H, Fujioka S, Takatsuto S, Yoshida S, An CS, Lee I, Hwang I, Choe S. (2006) The regulation of DWARF4 expression is likely a critical mechanism in maintaining the homeostasis of bioactive brassinosteroids in Arabidopsis. Plant Physiol 140: 548–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Hwang JY, Kim YS, Joo SH, Chang SC, Lee JS, Takatsuto S, Kim SK. (2005c) Arabidopsis CYP85A2, a cytochrome P450, mediates the Baeyer-Villiger oxidation of castasterone to brassinolide in brassinosteroid biosynthesis. Plant Cell 17: 2397–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolbe A, Schnerider B, Porzel A, Adam G. (1996) Metabolism of 24-epi-castasterone and 24-epi-brassinolide in cell suspension cultures of Ornithopus sativus. Phytochermistry 41: 163–167 [Google Scholar]
- Kolbe A, Schneider B, Porzel A, Schmidt J, Adam G. (1995) Acyl-conjugated metabolites of brassinosteroids in cell suspension cultures of Ornithopus sativus. Phytochemistry 38: 633–636 [Google Scholar]
- Kwon M, Choe S. (2005) Brassinosteroid biosynthesis and dwarf mutants. J Plant Biol 48: 1–15 [Google Scholar]
- Lee S, Lee DW, Lee Y, Mayer U, Stierhof YD, Lee S, Jürgens G, Hwang I. (2009) Heat shock protein cognate 70-4 and an E3 ubiquitin ligase, CHIP, mediate plastid-destined precursor degradation through the ubiquitin-26S proteasome system in Arabidopsis. Plant Cell 21: 3984–4001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Nagpal P, Vitart V, McMorris TC, Chory J. (1996) A role for brassinosteroids in light-dependent development of Arabidopsis. Science 272: 398–401 [DOI] [PubMed] [Google Scholar]
- Luo J, Fuell C, Parr A, Hill L, Bailey P, Elliott K, Fairhurst SA, Martin C, Michael AJ. (2009) A novel polyamine acyltransferase responsible for the accumulation of spermidine conjugates in Arabidopsis seed. Plant Cell 21: 318–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsolais F, Boyd J, Paredes Y, Schinas AM, Garcia M, Elzein S, Varin L. (2007) Molecular and biochemical characterization of two brassinosteroid sulfotransferases from Arabidopsis, AtST4a (At2g14920) and AtST1 (At2g03760). Planta 225: 1233–1244 [DOI] [PubMed] [Google Scholar]
- Mathur J, Molnár G, Fujioka S, Takatsuto S, Sakurai A, Yokota T, Adam G, Voigt B, Nagy F, Maas C, et al. (1998) Transcription of the Arabidopsis CPD gene, encoding a steroidogenic cytochrome P450, is negatively controlled by brassinosteroids. Plant J 14: 593–602 [DOI] [PubMed] [Google Scholar]
- Müssig C, Fischer S, Altmann T. (2002) Brassinosteroid-regulated gene expression. Plant Physiol 129: 1241–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Satoh T, Tanaka S, Mochizuki N, Yokota T, Nagatani A. (2005) Activation of the cytochrome P450 gene, CYP72C1, reduces the levels of active brassinosteroids in vivo. J Exp Bot 56: 833–840 [DOI] [PubMed] [Google Scholar]
- Nam KH, Dudareva N, Pichersky E. (1999) Characterization of benzylalcohol acetyltransferases in scented and non-scented Clarkia species. Plant Cell Physiol 40: 916–923 [DOI] [PubMed] [Google Scholar]
- Neff MM, Nguyen SM, Malancharuvil EJ, Fujioka S, Noguchi T, Seto H, Tsubuki M, Honda T, Takatsuto S, Yoshida S, et al. (1999) BAS1: a gene regulating brassinosteroid levels and light responsiveness in Arabidopsis. Proc Natl Acad Sci USA 96: 15316–15323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser JL, Maloof JN, Chory J. (2003) Building integrated models of plant growth and development. Plant Physiol 132: 436–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa N, Abe H, Natsume M, Shida A, Toyama S. (1995) Epimerization and conjugation of 14C-labeled in cucumber seedlings. J Plant Physiol 147: 294–300 [Google Scholar]
- Obayashi T, Kinoshita K, Nakai K, Shibaoka M, Hayashi S, Saeki M, Shibata D, Saito K, Ohta H. (2007) ATTED-II: a database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res (Database issue) 35: D863–D869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poppenberger B, Fujioka S, Soeno K, George GL, Vaistij FE, Hiranuma S, Seto H, Takatsuto S, Adam G, Yoshida S, et al. (2005) The UGT73C5 of Arabidopsis thaliana glucosylates brassinosteroids. Proc Natl Acad Sci USA 102: 15253–15258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouleau M, Marsolais F, Richard M, Nicolle L, Voigt B, Adam G, Varin L. (1999) Inactivation of brassinosteroid biological activity by a salicylate-inducible steroid sulfotransferase from Brassica napus. J Biol Chem 274: 20925–20930 [DOI] [PubMed] [Google Scholar]
- Shalit M, Guterman I, Volpin H, Bar E, Tamari T, Menda N, Adam Z, Zamir D, Vainstein A, Weiss D, et al. (2003) Volatile ester formation in roses. Identification of an acetyl-coenzyme A. Geraniol/Citronellol acetyltransferase in developing rose petals. Plant Physiol 131: 1868–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada Y, Fujioka S, Miyauchi N, Kushiro M, Takatsuto S, Nomura T, Yokota T, Kamiya Y, Bishop GJ, Yoshida S. (2001) Brassinosteroid-6-oxidases from Arabidopsis and tomato catalyze multiple C-6 oxidations in brassinosteroid biosynthesis. Plant Physiol 126: 770–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada Y, Goda H, Nakamura A, Takatsuto S, Fujioka S, Yoshida S. (2003) Organ-specific expression of brassinosteroid-biosynthetic genes and distribution of endogenous brassinosteroids in Arabidopsis. Plant Physiol 131: 287–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeno K, Kyokawa Y, Natsume M, Abe H. (2000) Teasterone-3-O-beta-D-glucopyranoside, a new conjugated brassinosteroid metabolite from lily cell suspension cultures and its identification in lily anthers. Biosci Biotechnol Biochem 64: 702–709 [DOI] [PubMed] [Google Scholar]
- Sohn EJ, Kim ES, Zhao M, Kim SJ, Kim H, Kim YW, Lee YJ, Hillmer S, Sohn U, Jiang L, et al. (2003) Rha1, an Arabidopsis Rab5 homolog, plays a critical role in the vacuolar trafficking of soluble cargo proteins. Plant Cell 15: 1057–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stündl U, Schneider B. (2001) 3Beta-brassinosteroid dehydrogenase activity in Arabidopsis and tomato. Phytochemistry 58: 989–994 [DOI] [PubMed] [Google Scholar]
- Suzuki H, Kim SK, Takahashi N, Yokota T. (1993) Metabolism of castasterone and brassinolide in mung bean explant. Phytochemistry 33: 1361–1367 [Google Scholar]
- Takahashi N, Nakazawa M, Shibata K, Yokota T, Ishikawa A, Suzuki K, Kawashima M, Ichikawa T, Shimada H, Matsui M. (2005) shk1-D, a dwarf Arabidopsis mutant caused by activation of the CYP72C1 gene, has altered brassinosteroid levels. Plant J 42: 13–22 [DOI] [PubMed] [Google Scholar]
- Tanaka K, Asami T, Yoshida S, Nakamura Y, Matsuo T, Okamoto S. (2005) Brassinosteroid homeostasis in Arabidopsis is ensured by feedback expressions of multiple genes involved in its metabolism. Plant Physiol 138: 1117–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton LE, Rupasinghe SG, Peng H, Schuler MA, Neff MM. (2010) Arabidopsis CYP72C1 is an atypical cytochrome P450 that inactivates brassinosteroids. Plant Mol Biol 74: 167–181 [DOI] [PubMed] [Google Scholar]
- Turk EM, Fujioka S, Seto H, Shimada Y, Takatsuto S, Yoshida S, Denzel MA, Torres QI, Neff MM. (2003) CYP72B1 inactivates brassinosteroid hormones: an intersection between photomorphogenesis and plant steroid signal transduction. Plant Physiol 133: 1643–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk EM, Fujioka S, Seto H, Shimada Y, Takatsuto S, Yoshida S, Wang H, Torres QI, Ward JM, Murthy G, et al. (2005) BAS1 and SOB7 act redundantly to modulate Arabidopsis photomorphogenesis via unique brassinosteroid inactivation mechanisms. Plant J 42: 23–34 [DOI] [PubMed] [Google Scholar]
- Voigt B, Porzel A, Naumann H, Hörhold-Schubert C, Adam G. (1993) Hydroxylation of the native brassinosteroids 24-epicastasterone and 24-epibrassinolide by the fungus Cunninghamella echinulata. Steroids 58: 320–323 [DOI] [PubMed] [Google Scholar]
- Walker K, Long R, Croteau R. (2002) The final acylation step in taxol biosynthesis: cloning of the taxoid C13-side-chain N-benzoyltransferase from Taxus. Proc Natl Acad Sci USA 99: 9166–9171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J. (2001) BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410: 380–383 [DOI] [PubMed] [Google Scholar]
- Watanabe T, Yokota T, Shibata K, Nomura T, Seto H, Takatsuto S. (2000) Cryptolide, a new brassinolide catabolite with a 23-oxo group from Japanese cedar pollen anther and its synthesis. J Chem Res 2000: 18–19 [Google Scholar]
- Weigel D, Ahn JH, Blázquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrándiz C, Kardailsky I, Malancharuvil EJ, Neff MM, et al. (2000) Activation tagging in Arabidopsis. Plant Physiol 122: 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto R, Fujioka S, Iwamoto K, Demura T, Takatsuto S, Yoshida S, Fukuda H. (2007) Co-regulation of brassinosteroid biosynthesis-related genes during xylem cell differentiation. Plant Cell Physiol 48: 74–83 [DOI] [PubMed] [Google Scholar]
- Yang Q, Reinhard K, Schiltz E, Matern U. (1997) Characterization and heterologous expression of hydroxycinnamoyl/benzoyl-CoA:anthranilate N-hydroxycinnamoyl/benzoyltransferase from elicited cell cultures of carnation, Dianthus caryophyllus L. Plant Mol Biol 35: 777–789 [DOI] [PubMed] [Google Scholar]
- Yokota T, Matsuoka T, Koarai T, Nakayama M. (1996) 2-Deoxybrassinolide, a brassinosteroid from Pisum sativum seed. Phytochemistry 42: 509–511 [Google Scholar]
- Yu XH, Gou JY, Liu CJ. (2009) BAHD superfamily of acyl-CoA dependent acyltransferases in Populus and Arabidopsis: bioinformatics and gene expression. Plant Mol Biol 70: 421–442 [DOI] [PubMed] [Google Scholar]
- Yuan T, Fujioka S, Takatsuto S, Matsumoto S, Gou X, He K, Russell SD, Li J. (2007) BEN1, a gene encoding a dihydroflavonol 4-reductase (DFR)-like protein, regulates the levels of brassinosteroids in Arabidopsis thaliana. Plant J 51: 220–233 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.