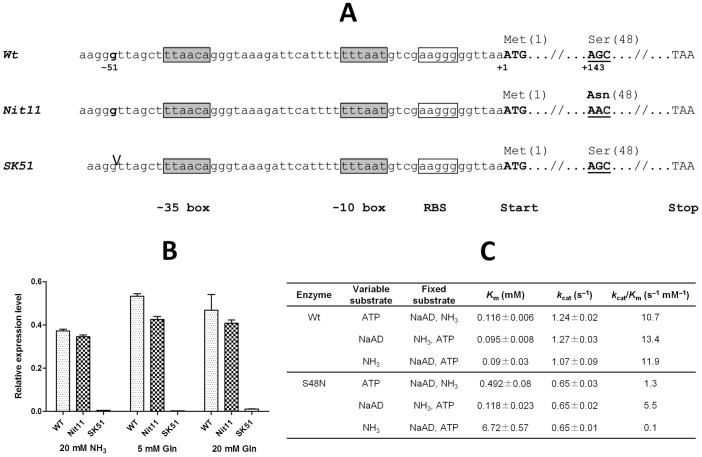
Figure 4. Analysis of Nit11 and SK51 S. typhimurium mutant strains.
(A) Schematic of S. typhimurium nadE mutant strains. Nit11 mutant features a missense mutation at nucleotide 143 yielding the amino acid substitution AN. SK51 mutant features a single nucleotide deletion (G at position – 51 considering as +1 the first base of the start codon) that is just upstream of the -35 box regulatory region of the promoter. Predicted regulatory sequences are indicated in the bottom line. (B) Relative expression level of nadE in the wild type and in the two classes of mutants nit11 (S48N) and SK51 in four different growth conditions: rich medium (1), minimal medium supplemented with 20 mM NH3 (2), MM supplemented with 5 mM (3) or 20 mM (4) glutamine. (C) Kinetic characterization of wild type and S48N S. typhimurium NAD synthetase. Initial rates were measured by spectrophotometrical coupled (SPEC) assays. The kinetic parameters K m and k cat are apparent values determined at fixed (saturating) concentrations of co-substrates. For fixed substrates, concentrations were: 2 mM ATP, 2 mM NaAD, and 40 mM NH3. Errors represent standard deviation.