Abstract
Aminoacyl-tRNA synthetases activate specific amino acid substrates and attach them via an ester linkage to cognate tRNA molecules. In addition to cognate proline, prolyl-tRNA synthetase (ProRS) can activate cysteine and alanine and misacylate tRNAPro. Editing of the misacylated aminoacyl-tRNA is required for error-free protein synthesis. An editing domain (INS) appended to bacterial ProRS selectively hydrolyzes Ala-tRNAPro, whereas Cys-tRNAPro is cleared by a freestanding editing domain, YbaK, through a unique mechanism involving substrate sulfhydryl chemistry. The detailed mechanism of catalysis by INS is currently unknown. To understand the alanine specificity and mechanism of catalysis by INS, we have explored several possible mechanisms of Ala-tRNAPro deacylation via hybrid QM/MM calculations. Experimental studies were also performed to test the role of several residues in the INS active site, as well as various substrate functional groups in catalysis. Our results support a critical role for the tRNA 2′-OH group in substrate binding and catalytic water activation. A role is also proposed for the protein’s conserved GXXXP loop in transition state stabilization and for the main chain atoms of Gly261 in a proton relay that contributes substantially to catalysis.
Keywords: Prolyl-tRNA synthetase, INS domain, aminoacyl-tRNA, post-transfer editing mechanism, QM/MM
INTRODUCTION
Aminoacyl-tRNA synthetases (aaRSs) play a critical role in protein synthesis. These enzymes catalyze the covalent attachment of amino acids onto their corresponding tRNAs.1 Catalysis by aaRSs follows a two-step mechanism wherein amino acids are first activated by ATP to form an aminoacyl-adenylate intermediate. In the second step, the activated amino acids are transferred to either the 2′-OH or the 3′-OH of the tRNA via a trans-esterification reaction. The product of this reaction, aminoacyl-tRNA, is then transported to the ribosomal machinery by elongation factors. AaRSs often attach structurally similar non-cognate amino acids to their corresponding tRNAs. If these mistakes are left uncorrected, errors in newly synthesized proteins accumulate and the resulting mutations can cause mis-folded or inactive proteins, leading to cytotoxicity.2 Several proofreading mechanisms have evolved to minimize such errors,3 including editing of aminoacyl-tRNAs either by domains appended to aaRSs in cis, or by free-standing editing domains that function in trans.4–7 Loss in editing function of aaRSs has been linked to disease phenotypes.8,9
AaRSs have been shown to carry out two types of editing. Misactivated aminoacyl-adenylates are cleared by “pre-transfer” editing, while mischarged aminoacyl-tRNAs are cleared via “post-transfer” editing.10,11 AaRSs are classified into two classes (class I and class II) based on the evolution of their catalytic domains (aminoacylation active site), and a subset of both classes of aaRSs encode post-transfer editing domains that are structurally and spatially distinct from the ancient catalytic core.
Escherichia coli prolyl-tRNA synthetase (ProRS), a class II synthetase, attaches non-cognate amino acids alanine and cysteine to its cognate tRNAPro at high enough frequencies to require editing. Ala-tRNAPro is cleared by an editing (INS) domain present in most bacterial ProRSs.12,13 In contrast, Cys-tRNAPro is not hydrolyzed by INS, but is cleared by a free-standing domain known as YbaK, which is homologous to the INS domain.4,5,14,15 Both INS and YbaK belong to the YbaK superfamily that also includes several other known or putative editing domains including YeaK, PrdX, ProX, and PA2301.4,15,16
Post-transfer editing involves cleavage of the ester bond between the amino acid and the 3′-terminal ribose moiety of the tRNA. However, the mechanism of catalysis differs for different editing domains. In the case of YbaK, the reaction is initiated by a nucleophilic attack of the sulfhydryl group of the substrate cysteine on the carbonyl carbon, leading to the formation of a thiolactone intermediate and concomitant cleavage of the ester bond.16 For the majority of editing domains, the reaction is likely initiated by nucleophilic attack of a strategically positioned water molecule; however, the mechanism of catalytic water activation, transition state stabilization, and proton transfer from the water to the 3′-terminal ribose on A76 of the tRNA may differ significantly between different systems. One recurring theme in editing mechanisms investigated to date, is the role of substrate 2′ or 3′-OH groups. For example, the crystal structure of a D-aminoacyl-tRNA deacylase-like domain in Pyrococcus abyssi threonyl-tRNA synthetase (ThrRS) suggests that the 2′-OH plays a role in activation of a catalytic water molecule for nucleophilic attack.17 Critical importance of the 3′-OH group of the substrate in editing activities has been previously shown in the case of both isoleucyl-tRNA synthetase (IleRS) and valyl-tRNA synthetase (ValRS).18 A role for the 3′-OH group in activation of the nucleophilic water molecule has also been implicated in the case of E. coli phenylalanyl-tRNA synthetase (PheRS)19 and recent quantum mechanical/molecular mechanical (QM/MM) studies of the Thermus thermophilus leucyl-tRNA synthetase (LeuRS) CP1 editing domain suggest a similar role for the 3′-OH in catalysis.20,21
The mechanism of hydrolysis by the INS domain is poorly understood, in part, due to the lack of high-resolution structural data on its mode of substrate binding. We have recently reported a computational model of the INS domain bound to a Ala-tRNAPro substrate analog, 5′-CCA-Ala.22 This model together with biochemical data allowed us to propose a binding site for the substrate alanine in which the methyl side chain fits into a well-defined and tunable hydrophobic pocket. However, the exact orientation of the substrate in the catalytic center and the detailed mechanism of hydrolysis remain incompletely understood. Interestingly, the INS active site is notable for its lack of conserved residues whose side chains may play a role in catalysis.
In this study, we have combined hybrid QM/MM calculations with biochemical assays to probe the role of various substrate functional groups and putative active site residues in catalysis by the E. coli ProRS INS domain. Guided by the requirement of several key mechanistic steps, such as activation of a catalytic water and stabilization of the transition state, we explored several feasible computational mechanisms for catalysis. These mechanisms were evaluated experimentally by altering substrate functional groups and amino acid residues that were predicted to play a role in catalysis.
Our results are consistent with a mechanism wherein hydrolysis of Ala-tRNAPro by INS is initiated via activation of a catalytic water molecule by the 2′-OH of the A76 ribose of tRNA. The tetrahedral intermediate formed is stabilized by the backbone amides of the conserved 331GXXXP loop. A proton from the catalytic water is shuttled to the O3′ of the tRNA via the backbone carbonyl of the Gly261 residue. Ser280 and Glu265 residues play important roles in stabilization of the reaction intermediates. Consistent with this mechanism, substitution of the 2′-OH of the substrate or deletion of Gly261 leads to complete loss of hydrolysis activity, whereas mutation of Ser280 or Glu265 leads to significant loss in activity. These results support the role of both substrate functional groups and the protein backbone in catalysis.
METHODS
Computational methods
The structural model of the Enterococcus faecalis ProRS INS domain bound to the Ala-tRNAPro analog 5′-CCA-Ala22 was used as a starting structure for the hybrid QM/MM calculations. The initial INS domain:CCA-Ala complex including important structural and catalytic water molecules in the active site of INS domain, obtained from a 10 ns MD simulation of INS:CCA-Ala complex in a solvated shell at 300 K, was solvated in an 8 Å octahedral shell of TIP3P23 waters and optimized using the ff0324 force field in AMBER10.25 The structure was further optimized using the hybrid QM/MM method as described below.
Hybrid QM/MM calculations
All QM/MM optimizations were carried out using ChemShell26,27 as an interface to TURBOMOLE (v5.10)28 for the QM atoms, and ChemShell’s internal version of DL_POLY29 for the MM treatment using the CHARMM force field.30 Geometry optimization was carried out using the DL-FIND optimizer, employing the Broyden–Fletcher–Goldfarb–Shanno (BFGS) method. The QM layer was treated using density functional theory (DFT), specifically the BP8631,32 functional with the SV(P)33 basis set, and single-point energy calculations were carried out for each optimized geometry with the same functional and the TZVPP34 basis set. QM atoms included the substrate alanine, the ribose moiety of A76 of the tRNA analog, Ser280 and His366 side chains, 331GXXXP backbone atoms, and several structural and catalytic water molecules. In addition, the A76 base of the tRNA and backbone atoms of several additional loops in the vicinity of the substrate were included, depending on the mechanism being examined (Supplementary Figure 1). Typically, 90–95 atoms were included in the QM layer. All residues with any atom within 8Å of the substrate alanine were optimized with MM treatment, while the remaining residues were fixed. The QM and MM regions were coupled via electrostatic embedding, with electrostatic interactions handled by the QM code with QM polarization. Potential energy surface scans were carried out by varying the reaction coordinate, defined as the difference between the Ala-C(O)–O3′-A76 (bond cleaved) and Ala-C(O) –OH2 (bond formed), and optimizing all other degrees of freedom at each step of the fixed reaction coordinate.
Materials
All amino acids, 3-deaza-adenosine, and standard chemicals were purchased from Sigma unless otherwise noted. [3H]Alanine (54 Ci/mmol) and [14C]alanine (160 Ci/mmol) were from PerkinElmer. 2′-Fluoro-2′-deoxyadenosine-5′-triphosphate (2′-F-dATP) and 2′-Amino-2′-deoxyadenosine-5′-triphosphate (2′-NH2-dATP) were from Trilink Biotechnologies.
Protein expression and purification
Histidine-tagged wild-type (WT) E. coli ProRS,13 WT E. coli alanyl-tRNA synthetase (AlaRS),35 and E. coli tRNA nucleotidyl transferase (NTase)18 were purified using the His-select nickel affinity gel (Sigma) as previously described. The substitution and deletion mutants of E. coli ProRS were generated using the QuikChange site-directed mutagenesis kit (Stratagene). Mutations were confirmed by DNA sequencing (Genewiz). All E. coli ProRS variants were overexpressed in BL21(DE3) cells and purified as described for the WT protein, except that the deletion mutants were induced with 75 μM of isopropyl β-D-1-thiogalactopyranoside at room temperature for 12 h. The concentrations of E. coli AlaRS and NTase enzymes were determined by the Bradford assay.36 The concentrations of WT and mutant E. coli ProRSs were determined by active site titration.37
Synthesis of 3-deaza-ATP
3-deaza-ATP was synthesized from 3-deaza-adenosine as previously described.38 Briefly, 10 mg of 3-deaza-adenosine in 1 ml of cold trimethylphosphate (PO(OMe)3) was reacted with 2.9 equivalents of phosphorus oxychloride (POCl3, 99.999%) on ice overnight with constant stirring to form the monophosphate intermediate. To form the triphosphate, the reaction was then warmed to room temperature and was further reacted with 84 mg (4.1 equivalents) of tributylammonium pyrophosphate dissolved in 300 μl dry N,N-dimethylformamide (DMF) for 45 min at room temperature. The reaction was quenched with 5 ml of 0.1 M triethylammonium bicarbonate (TEAB). The solution was then diluted to ~200 ml with milliQ water and loaded onto a Sephadex-DEAE A25 ion-exchange column pre-equilibrated with 2.5 mM TEAB. Samples were eluted with a TEAB gradient (0 to 1 M). The presence of the 3-deaza-ATP product in the eluted fractions was evaluated by measuring the absorbance at 264 nm. The appropriate fractions were pooled, dried under vacuum (with the aid of EtOH), and dissolved in 0.1 M triethylammonium acetate (TEAA) (pH 7.0). The product was further purified by high-performance liquid chromatography using a C-18 column (GE Healthcare) and eluted with a 50-min gradient from 0 to 60% acetonitrile in 0.1 M TEAA. Final fractions containing 3-deaza-ATP were pooled, dried under vacuum and resuspended in milliQ water. The purity and identity of the product was verified by electrospray ionization mass spectrometry.
Preparation of tRNAs
E. coli tRNAPro (G1:C72,U70 variant to facilitate charging by AlaRS)39 was prepared by in vitro transcription as described.12 To prepare 3′-end modified tRNAs, a 75-mer run-off transcript lacking the 3′-terminal adenosine was generated by mutating the BstNI restriction site, CCAGG, to a SacII restriction site, CCGCGG, at the 3′-end of the tRNA gene in the DNA plasmid encoding the E. coli tRNAPro G1:C72,U70 variant. The resulting plasmid was treated with SacII restriction endonuclease for 16 h at 37 °C followed by in vitro transcription with T7 RNA polymerase. Full-length WT or modified tRNAs were generated by incubating 30 μM 75-mer tRNA with 10 μM E. coli NTase and 5 mM ATP (WT), 2′-deoxyATP (2′-dATP), 3-deaza-ATP, 2′-F-dATP, or 2′-NH2-dATP for 4 h at 37 °C. The reactions were worked up as previously described.40 Conversion to full-length 76-mer tRNAs was confirmed by analysis on 0.4 mm thick denaturing 12% polyacrylamide gel.
Preparation of aminoacyl-tRNA substrates
[3H]Ala-A76-tRNAPro (WT), [3H]Ala-2′-dA76-tRNAPro, [3H]Ala-2′-F-dA76-tRNAPro, [3H]Ala-2′-NH2-dA76-tRNAPro, and [3H]Ala-3deaza-A76-tRNAPro were prepared by incubating 8 μM of the respective tRNAs with 4 μM WT E. coli AlaRS and [3H]alanine (12.9 μM) in buffer containing 50 mM HEPES (pH 7.5), 4 mM ATP, 20 mM KCl, 20 mM β-mercaptoethanol, 25 mM MgCl2, and 0.1 mg/ml BSA for 4 h at 25 °C. Ala-tRNAPro variants (WT and 3′-end modified) and [14C]Ala-tRNAPro were similarly prepared using ~330 μM of unlabeled or [14C]-labeled alanine, respectively. Following aminoacylation reactions, the substrate aminoacyl-tRNAs were phenol-chloroform extracted, ethanol precipitated, resuspended in 50 mM KPO4 (pH 5.0) and stored at −20 °C for use in deacylation assays.
Deacylation assays
Deacylation of [3H]-Ala-tRNAPro (WT and 3′-end-modified) or [14C]Ala-tRNAPro was carried out according to published protocols.14,16 Reactions were performed using ~1 μM aminoacyl-tRNAs and 1.5–5 μM WT E. coli or mutant ProRS in buffer containing 150 mM KPO4 (pH 7.0), 5 mM MgCl2, 0.1 mg/ml BSA, and 15 μg/ml of inorganic pyrophosphatase (Roche) at 25 °C unless otherwise indicated. A background reaction lacking enzyme was carried out in each case. The reactions were monitored by precipitating the tRNA on Whatman 3MM filter pads followed by scintillation counting.5 All assays were carried out in triplicate, except for [3H]-Ala-2′-NH2-dA76-tRNAPro, which was performed only once for technical reasons. For competition assays, at least 10 μM of Ala-2′-dA76-tRNAPro or Ala-2′-F-dA76-tRNAPro were mixed with ~1 μM [14C]Ala-tRNAPro in similar reaction buffer, and reactions were initiated by addition of 1.5 μM WT ProRS at room temperature. In competition assays using the ProRS deletion mutants, 1.5 μM of WT ProRS was mixed with 9 μM of each of the deletion mutants in deacylation buffer, and reactions were initiated by the addition of [14C]Ala-tRNAPro at room temperature. Reactions were monitored by precipitation on filter pads as described above. The fraction of aminoacyl-tRNA remaining was plotted as a function of time following subtraction of the buffer-only background reaction, and fitted to a single-exponential equation to obtain kobs. All assays were carried out in triplicate.
RESULTS AND DISCUSSION
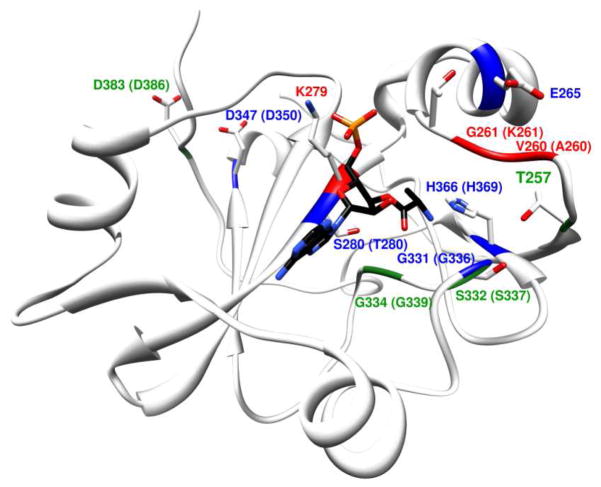
The bacterial ProRS INS domain has been previously investigated using biochemical,13,22,41 structural,42 and computational studies.22 Although there is no available crystal structure of an INS domain-tRNA complex, the computational model of E. faecalis INS bound to 5′-CCA-Ala,22 revealed that the aminoacyl moiety binds in a well-defined hydrophobic pocket formed by residues Ile263, Val266, Ile277, and Lys279 and rationalizes the high specificity of INS for the substrate alanine. This model was further supported by experimental mutagenesis studies, which showed a complete switch of the substrate specificity of the INS domain from alanine to either cysteine or serine, by tuning the size of this hydrophobic pocket. Thus, this model provides a foundation for further mechanistic studies. Interestingly, within the putative INS active site pocket, there are very few amino acid side chains that would be predicted to participate in catalysis. In fact, nearly all of the charged or polar residues in the active site have been investigated by mutagenesis13,41 (Figure 1), and the only residues with a significant impact on hydrolytic activity are Lys279, Gly331, and His366. Lys279 is conserved throughout the YbaK superfamily and is believed to bind the A76 phosphate of the tRNA, stabilizing the tRNA conformation.16,22 Gly331 is also conserved throughout the YbaK superfamily and is part of a conserved GXXXP motif, which may play an equivalent role to the oxyanionic hole in serine proteases43 and D-Tyr-tRNATyr deacylases.44 Thus, the Gly331 residue is likely involved in the stabilization of the oxyanionic transition state formed during the hydrolytic reaction. Based on modeling, His366 is involved in binding to the substrate through a H-bond to the amide group of the alanine moiety.22 Consistent with this proposal, an H366A ProRS variant has an altered deacylation substrate specificity from Ala-tRNA to Pro-tRNA.13 Two other residues, Asp347 and Asp383, show 2- to 5-fold decreases in deacylation activity upon mutation, but they lie outside the active-site pocket and most likely play structural roles.
Figure 1. Structural model of E. faecalis ProRS INS domain bound to Ala-tRNAPro substrate analog, 5′-CCA-Ala.
Active site residues investigated by mutagenesis in this study (V260, G261, E265, and S280) or in previous studies (T257, K279, G331, S332, G334, and H366 in E. faecalis ProRS41 and T257, K279, D350, and D386 in E. coli ProRS13) are depicted as stick models. The corresponding residues in E. coli ProRS are shown in parentheses in cases where they differ. Residues are colored based on the relative deacylation activity (kobs) of the mutants relative to WT. Green: ≤ 4-fold decrease; Blue: 4 – 40 fold decrease; Red: > 40-fold decrease.
Hydrolysis of the ester bond between the mis-charged amino acid and the tRNA requires several catalytic steps. In the first step, a water molecule must be activated by a base for nucleophilic attack of the carbonyl carbon of the scissile ester bond. The resulting oxyanionic transition state needs to be stabilized for effective catalysis. Finally, a proton is transferred from the nucleophilic water to the recently cleaved O3′ of the tRNA. Based on our model of INS:CCA-Ala, we can envision at least three unique scenarios for general base catalysis that are allowed by the geometry of the substrate in the active site pocket. Because of its proximity to the catalytic center, a likely candidate for the general base is the 2′-OH group of the 3′-terminal ribose of the substrate. Another candidate for this role is the N3 of the 3′-terminal adenine base of tRNA, which could abstract a proton from the catalytic water via the 2′-OH of the ribose. Finally, the backbone carbonyl of Gly261, which according to our model lies close to both the catalytic water and the O3′ of A76, may play a role in shuttling a proton from the water to the O3′. To establish which of these proposed mechanisms is most feasible, we computed the potential energy surface of each proposed mechanism and experimentally probed functional groups of the substrate, as well as the enzyme, to corroborate the findings from the theoretical investigation.
Potential energy surface scans of mechanisms of hydrolysis
Potential energy surface scans were performed using a hybrid QM/MM approach wherein the catalytic center, such as the substrate alanine, the terminal ribose of the tRNA, side chains of His366 and Ser280, backbone atoms of the GXXXP loop, and catalytic and structural waters were treated with DFT methods, and the surrounding protein and solvent molecules were treated with a force field as described in the methods section. To calculate the potential energy surface, reaction geometries were optimized at 0.2 Å steps along the reaction coordinate using the bond length difference constraint, while the reactant and product geometries were optimized without any constraints. The reaction coordinate refers to the difference between the bond to be cleaved (Ala-C(O) – O3′-tRNA) and the bond formed (Ala-C(O) – OH2). It should be noted that the calculations described here ignore entropic changes in the system and thus the calculated energy barriers represent total energy differences rather than the free energy differences. The entropic contributions to the different mechanistic options would be expected to be relatively similar, leading us to a meaningful conclusion about the best mechanistic pathway from the calculated energies in this system. This approach provides an approximation of the activation barriers; however, the relative activation barriers for different mechanisms can be estimated to a high degree.
Mechanism of hydrolysis with N3 of A76 adenine as general base
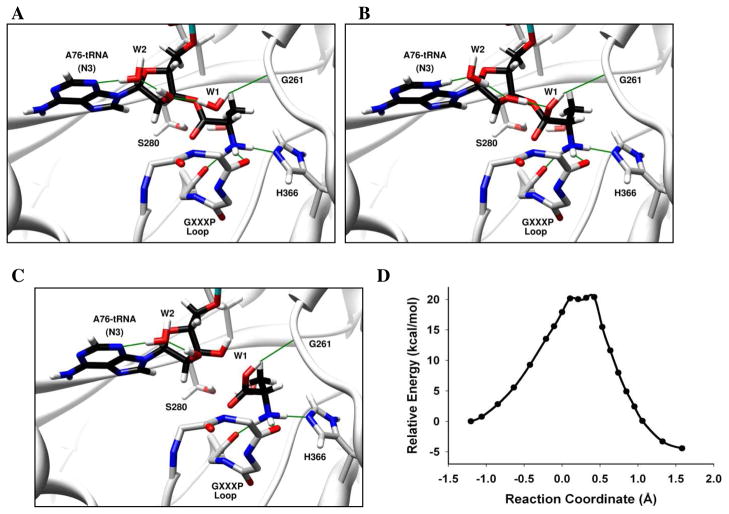
Several studies of ribozymes, including Hepatitis Delta Virus ribozyme, hammerhead ribozyme, and Varkud satellite ribozyme, have demonstrated the role of nucleobases in general acid-base catalysis.45–47 In our model of the INS domain bound to 5′-CCA-Ala, the N3 atom of A76 lies close to the 2′-OH of the substrate. We evaluated the possibility that A76 activates the catalytic water molecule by hybrid QM/MM calculations (Figure 2). In the reactant geometry (Figure 2A), the catalytic water (W1) forms a H-bond with the 2′-OH of the substrate, which in turn H-bonds with another water (W2) that is H-bonded to N3 of A76. As the reaction progresses, a proton from W1 is shuttled to N3 via the 2′-OH and W2, via a Grotthuss structural diffusion,48 leading to a protonated A76 intermediate (Figure 2B). In the next step, the proton from the protonated A76 is shuttled back to O3′ of the tRNA via W2 and 2′-OH, forming a stable hydrolyzed product (Figure 2C). The energy barrier calculated for this mechanism is ~ 20 kcal/mol (Figure 2D). We also examined a similar mechanism with A76 mutated to 3-deaza-A, wherein the N3 atom of A76 is replaced with CH. In this case, a proton is transferred from W1 to the 2′-OH, but downstream transfer to A76 is not feasible. The barrier for this mechanism is also ~ 20 kcal/mol (data not shown), suggesting that protonation of the 2′-OH is the rate-limiting step. Consistent with a relatively high energy barrier for this mechanism, experimental deacylation assays performed with E. coli ProRS and an Ala-3-deaza-A76-tRNAPro substrate did not show any significant loss in activity compared to the WT Ala-tRNAPro substrate (Figure 3A). Overall, these results suggest that a mechanism involving A76 as a general base, while theoretically feasible, is unlikely to be the mechanism for INS-catalyzed Ala-tRNAPro hydrolysis.
Figure 2. Deacylation of Ala-tRNAPro by E. faecalis ProRS INS domain via A76 as general base.
(A) Catalytic water (W1) is activated by the 2′-OH. (B) A proton transfer occurs from W1 to N3 of A76 via the 2′-OH and W2 to form a protonated A76 intermediate. (C) A proton is subsequently transferred to O3′ via W2 and 2′-OH to form the hydrolyzed product. The oxyanionic intermediate is stabilized by the GXXXP loop. The reaction coordinate refers to bond broken (Ala-C(O) – O3′-A76) – bond formed (Ala-C(O) – OH2). Panels A, B and C show the optimized reactant, intermediate, and products, respectively. (D) The calculated potential energy surface for this mechanism at the BP86/TZVPP//BP86/SV(P) level of theory for the QM domain.
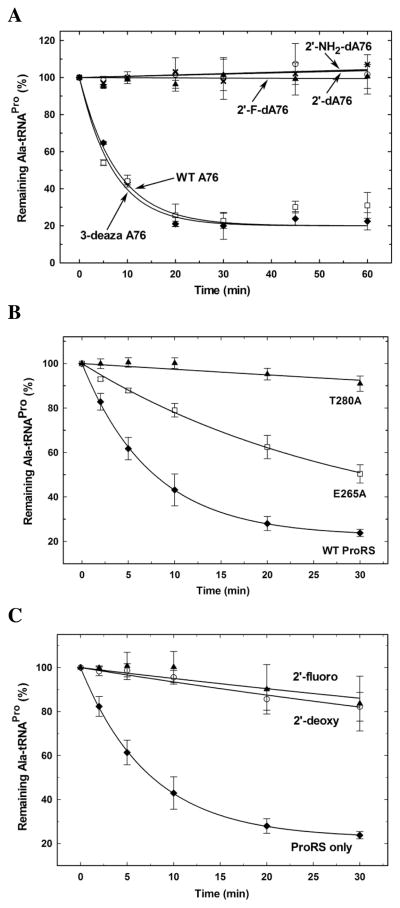
Figure 3. Effect of mutations in substrate functional groups and active site residues on Ala-tRNAPro hydrolysis by E. coli ProRS.
(A) Deacylation of WT [3H]Ala-tRNA and 3′-end modified variants (WT A76 (◆), 3deaza-A76 (□), 2′-dA76 (○), 2′-NH2-dA76 (✕) and 2′-F-dA76 (▲)) with 5 μM of WT E. coli ProRS in deacylation buffer without inorganic pyrophosphatase. (B) Effect of point mutations in the INS active site. Deacylation of [14C]Ala-tRNAPro by 1.5 μM WT (◆), E265A (□), and T280A (▲) E. coli ProRS. (C) Competitive inhibition assays to probe binding of 3′-end modified Ala-tRNAPro substrate analogs. Deacylation of [14C]Ala-tRNAPro by 1.5 μM WT E. coli ProRS in the absence (ProRS only, ◆) and presence of ~10 μM Ala-2′-dA76-tRNAPro (2′-deoxy, ○) and Ala-2′F-dA76-tRNAPro (2′-fluoro, ▲).
Water activation and proton transfer by 2′-OH
QM/MM studies of the LeuRS CP1 editing domain suggest that substrate hydrolysis proceeds via activation of the catalytic water by the 3′-OH and transfer of a proton from the catalytic water to the O2′ of the cleaved ester bond.21 The energy barrier reported for this mechanism is ~20 kcal/mol. The 3′-endo sugar pucker of the A76 ribose and the orientation of the substrate carbonyl in our model are not consistent with such a proton transfer; however, a rotation of the docked 5′-CCA-Ala substrate along the scissile ester bond would allow concerted proton transfer from the catalytic water (W1) to the O3′ via the 2′-OH group. Subsequent formation of a H-bond between the substrate carbonyl and the Ser280 hydroxyl could stabilize the oxyanionic transition state (Figure 4A–C). A potential energy surface scan of this mechanism results in a theoretical energy barrier of 17 kcal/mol (Figure 4D). Since Ser280 plays a critical role in this mechanism, we performed Ala-tRNAPro deacylation assays with a T280A E. coli ProRS variant (Thr280 in E. coli corresponds to Ser280 in E. faecalis). This substitution results in a 40-fold loss in enzymatic activity (Figure 3B), supporting this as a feasible mechanism of hydrolysis by the INS domain.
Figure 4. Deacylation of Ala-tRNAPro by E. faecalis ProRS INS domain via 2′-OH as general base.
(A) The catalytic water (W1) is activated by the 2′-OH. (B) A proton from W1 is transferred to O3′ via the 2′-OH in a concerted mechanism. (C) Oxyanionic intermediate is stabilized by the S280 sidechain. The reaction coordinate refers to bond broken (Ala-C(O) – O3′-A76) – bond formed (Ala-C(O) – OH2). Panels A, B and C show the optimized reactant, transition state, and products, respectively. (D) The calculated potential energy surface for this mechanism at the BP86/TZVPP//BP86/SV(P) level of theory for the QM domain.
Water activation by 2′-OH and proton transfer by Gly261 backbone carbonyl
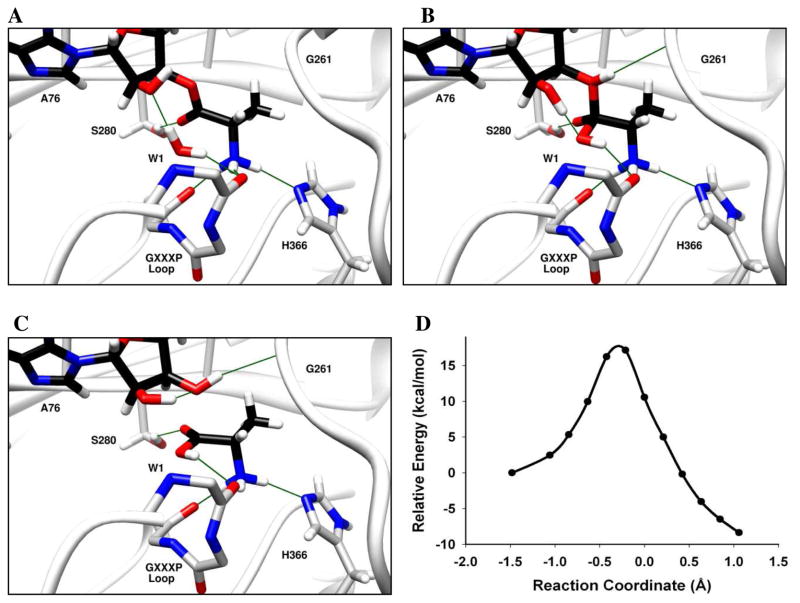
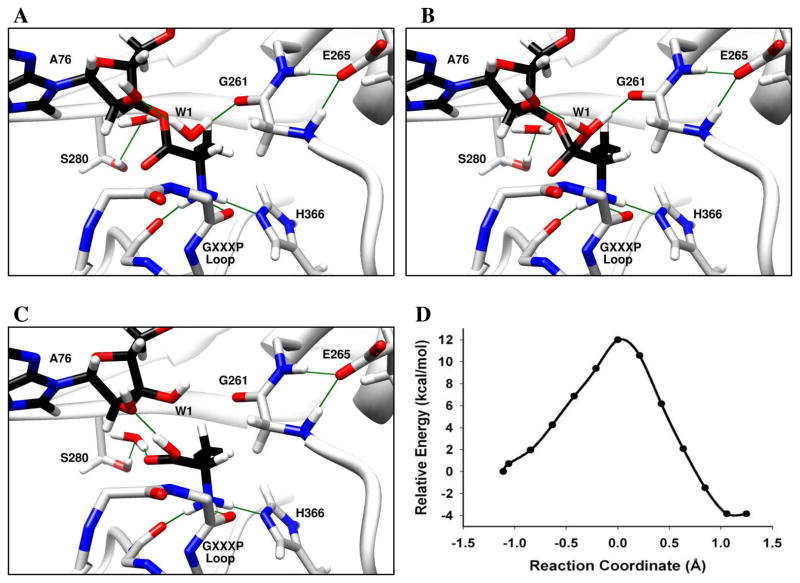
Backbone atoms within catalytic centers are often involved in key H-bonding interactions that contribute to catalysis.49–51 Due to the high polarizability of the amide bond, backbone atoms may also participate directly in catalytic processes such as proton transfer. For example, QM/MM simulations of aldehyde hydrogenase suggest that a backbone amide proton is transferred to the reaction intermediate during catalysis.51 In the computational model of INS:CCA-Ala, the backbone carbonyl of Gly261 forms a low barrier hydrogen bond52,53 with the catalytic water and is situated in an ideal location for proton transfer from the catalytic water to the O3′ of the tRNA. Also, the backbone amides of Gly261 and Thr262 form H-bonds with the Glu265 side chain, which results in an increased electron density on the carbonyl oxygen of Gly261. We calculated the potential energy surface of a mechanism wherein the catalytic water (W1) is activated by the 2′-OH of the substrate (Figure 5). As the reaction progresses along the reaction coordinate, the bond between the oxygen atom of W1 and the carbonyl carbon of the substrate is formed, and a proton from W1 is transiently shared with the Gly261 carbonyl (Figure 5A) in a Zundel-like complex.54 After the reaction crosses the transition state, the bond between O3′ of tRNA and the carbonyl carbon elongates, and a proton is shared between the Gly261 carbonyl and the O3′ of the tRNA (Figure 5B). This proton transfer is assisted by quantum fluctuations of the structural defect represented by the protonated carbonyl of Gly261 residue, and is stabilized by the formation of a barrier-less Zundel-like complex where the proton is shared by oxygen atoms of Gly261 carbonyl and O3′ of the tRNA. Cleavage of the ester bond is accompanied by transfer of the shared proton to O3′, leading to the formation of a stable product (Figure 5C). The oxyanionic transition state is stabilized by backbone amides of the 331GXXXP loop. The energy barrier for this mechanism was calculated to be 12 kcal/mol, making it the most facile mechanism investigated in this study (Figure 5D).
Figure 5. Deacylation of Ala-tRNAPro by E. faecalis ProRS via proton relay involving Gly261 backbone carbonyl.
(A) Catalytic water (W1) is activated by the 2′-OH. (B) Proton transfer from W1 to O3′ is facilitated by the Gly261 backbone carbonyl. The oxyanionic intermediate is stabilized by the GXXXP loop. (C) Transfer of the proton to O3′ and formation of product. The reaction coordinate refers to bond broken (Ala-C(O) – O3′-A76) – bond formed (Ala-C(O) – OH2). Panels A, B and C show the optimized reactant, transition state, and products, respectively. (D) The calculated potential energy surface for this mechanism at the BP86/TZVPP//BP86/SV(P) level of theory for the QM domain.
In support of this mechanism, substitution of a 2′-deoxy, 2′-fluoro, or 2′-amino sugar at A76 resulted in a complete loss of detectable deacylation activity (Figure 3A). On the basis of these results, we estimate that these single functional group substitutions of the 2′-OH result in at least a 1000-fold decrease in deacylation efficiency. We also show that Ala-2′-dA76-tRNAPro or Ala-2′-F-dA76-tRNAPro can competitively inhibit Ala-tRNAPro deacylation by E. coli ProRS (Figure 3C), which indicates that binding is not significantly affected by these substitutions. These results are consistent with a role for the 2′-OH group in activation of the catalytic water. Alternatively, the mechanism for hydrolysis may require discharge from the 2′-OH group. However, our previously published model of E. faecalis INS bound to 5′-CCA-Ala,22 is not consistent with the tRNA bound with O2′-Ala connectivity, which would result in significant steric clash of the substrate ribose or the terminal adenine base with the protein (data not shown). Mutagenesis studies provide additional support for the proposed mechanism. Alanine substitution of Gly331, part of GXXXP loop proposed to be required for stabilization of the transition state, has previously been shown to result in a 6-fold loss in deacylation activity.41 We also show that E265A E. coli ProRS is ~ 4-fold reduced in activity relative to the WT enzyme (Figure 3B). The Ser280 hydroxyl group may also participate in this mechanism by stabilizing the O3′ of A76 via a structural water molecule (Figure 5). Thus, a 40-fold loss in activity of T280A E. coli ProRS is also consistent with this mechanism.
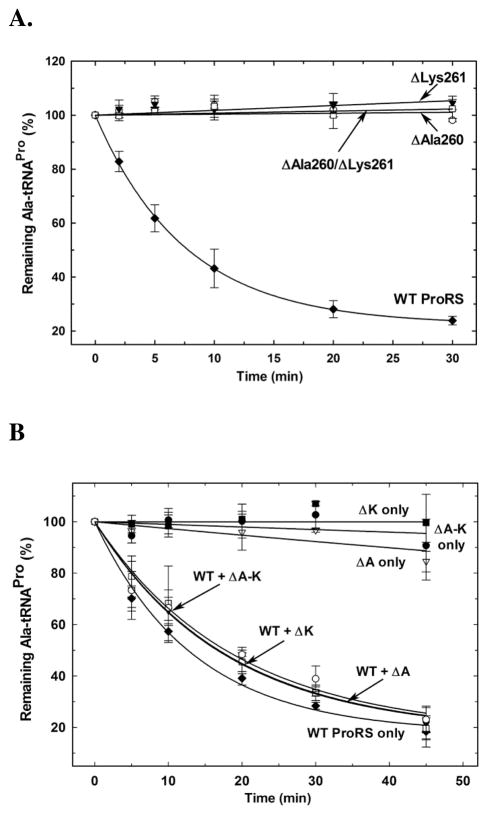
The mechanism described above suggests a critical role for the Gly261 backbone carbonyl in catalysis. The carbonyl backbone of the neighboring residue Val260 also points towards the substrate binding pocket of the INS domain. Neither of these residues is conserved in bacterial ProRSs. To experimentally examine their role in catalysis, substitution mutations of the residues are not informative, since their side chains point away from the active site of the INS domain and are not involved in catalysis. Thus, we tested single and double deletion mutants of the corresponding residues in E. coli ProRS (ΔAla260, ΔLys261, and ΔAla260/ΔLys261). All three mutants examined showed a >1000-fold loss in Ala-tRNAPro deacylation activity (Figure 6A), consistent with our hypothesis that the backbone atoms of these residues play an active role in catalysis. Deletion of single residues from the substrate binding loop may also affect binding of the substrate. To investigate this possibility, we performed Ala-tRNAPro deacylation assays by E. coli ProRS in the presence of each of the deletion mutants. The lack of competitive inhibition by even a 6-fold excess of the ProRS deletion mutants (Figure 6B) indicated a defect in tRNA substrate binding ability. Although substrate binding is affected, none of the deletion mutants were defective in their cognate proline charging ability relative to WT ProRS (data not shown). We also found that the global fold of each of these deletion mutants is similar to that of the WT protein, as evaluated by circular dichroism (CD) spectroscopy (Supplementary Figure 2). Thus, although Ala-tRNAPro substrate binding appears to be somewhat compromised, the complete loss in deacylation activity of the mutants is consistent with a functional role for the Gly261 backbone carbonyl in catalysis.
Figure 6. Effect of deletion of active site residues on Ala-tRNAPro hydrolysis by E. coli ProRS.
(A) [14C]Ala-tRNAPro deacylation by 1.5 μM WT (◆), ΔAla260 (○), ΔLys261 (▼), and ΔAla260/ΔLys261 (□) E. coli ProRS. (B) [14C]Ala-tRNAPro deacylation by 1.5 μM E. coli ProRS in the absence (WT ProRS only, ◆) or presence of ~9 μM ΔAla260 (WT+ ΔA, ▼), ΔLys261 (WT+ ΔK, □), and ΔAla260/ΔLys261 (WT+ ΔA-K, ○) ProRS. Deacylation by ~9 μM ΔAla260 (ΔA only, ▽), ΔLys261 (ΔK only, ■), and ΔAla260/ΔLys261 (ΔA-K only, ●) ProRS is also shown.
Role of protein main chain carbonyl groups in catalysis in similar systems
Our QM/MM calculations suggest that the dominant mechanism of catalysis by the INS domain involves the activation of a catalytic water by the substrate 2′-OH and proposes a role for the Gly261 backbone in proton shuttling. This mechanism of catalysis may also be common to some other deacylases. In YbaK, the substrate binds in an orientation similar to that observed in the model of the INS domain.16 YbaK, which is a structural homolog of INS, also lacks catalytic protein side chains in its active site and the 2′-OH group of the substrate A76 is also required for catalysis.16 Thus, although the substrate sulfhydryl acts as the nucleophile in this case, these observations suggest that YbaK may also share some features of the proposed INS mechanism of catalysis. Another example is the structurally distinct editing domain of ThrRS from P. abyssi (Pab-NTD). The crystal structure of Pab-NTD bound to post-transfer editing substrate analog 3′-(L-seryl) amino-3′-deoxyadenosine (Ser3AA) shows a lack of direct involvement of protein side chains in post-transfer editing.17 It was hypothesized that the 2′-OH of the substrate A76, which is the only base in the vicinity of the labile bond, may play a role in activation of catalytic water. A closer examination of this structure shows that the backbone carbonyl of Tyr119 has a direct interaction with the catalytic water, as well as the nitrogen atom of the amide linkage of the substrate analog, which would correspond to the O3′ atom of the unmodified substrate. The lack of any other protein or substrate functional group in the vicinity of the scissile bond suggests that the Tyr119 backbone carbonyl may play a role in proton shuttling in Pab-NTD.
CONCLUSIONS
We have previously characterized the substrate binding pocket of the E. faecalis ProRS INS domain by computational and experimental methods.22 This model showed a clear lack of direct interactions of protein side chains with the substrate in a way that could facilitate catalysis. In this report, our extensive QM/MM calculations show that catalysis by INS may not require active participation of protein side chains. The most feasible mechanism of catalysis involves the activation of the catalytic water by the substrate 2′-OH group and a proton shuttle from a catalytic water to the nascent O3′ of the tRNA via the backbone carbonyl of the non-conserved Gly261 residue. We also confirm experimentally that the 2′-OH is critical for catalysis by E. coli ProRS, but not for substrate binding. Overall, this mechanism of catalysis resembles the “hybrid ribozyme/protein catalysis” mechanism proposed for post-transfer editing by the LeuRS editing domain.21 However, in the INS domain, in addition to the substrate 2′-OH group, functional groups of the protein’s backbone play an active role in catalysis. The Gly261 carbonyl facilitates a critical proton transfer and backbone amides of the GXXXP loop help to stabilize the transition state. The Glu265 residue stabilizes the loop containing the Gly261 residue, and the Ser280 residue stabilizes the O3′ of the scissile ester bond. Our findings support the role of protein backbone atoms in catalysis and underscore the importance of including main chain atoms in QM or QM/MM investigations of enzyme mechanisms.
Supplementary Material
Acknowledgments
We thank Prof. P. Schimmel (Scripps Research Institute, CA) for the plasmid encoding E. coli tRNA nucleotidyl transferase. Generous computational resources were provided by the Ohio Supercomputer Center. This work was funded by NIH grant GM049928 (to K.M-F.).
Footnotes
ASSOCIATED CONTENT
Supporting Information Available. Structure of QM atoms used in hybrid QM/MM calculations and folding studies of deletion mutants of E. coli ProRS. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Ibba M, Söll D. Annu Rev Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 2.Reynolds NM, Lazazzera BA, Ibba M. Nat Rev Microbiol. 2010;8:849–856. doi: 10.1038/nrmicro2472. [DOI] [PubMed] [Google Scholar]
- 3.Mascarenhas AP, An S, Rosen AE, Martinis SA, Musier-Forsyth K. Protein Eng. Vol. 22. Springer; Berlin Heidelberg: 2009. pp. 155–203. [Google Scholar]
- 4.Ahel I, Korencic D, Ibba M, Söll D. Proc Natl Acad Sci USA. 2003;100:15422–15427. doi: 10.1073/pnas.2136934100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.An S, Musier-Forsyth K. J Biol Chem. 2004;279:42359–42362. doi: 10.1074/jbc.C400304200. [DOI] [PubMed] [Google Scholar]
- 6.Chong YE, Yang XL, Schimmel P. J Biol Chem. 2008;283:30073–30078. doi: 10.1074/jbc.M805943200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ling J, Reynolds N, Ibba M. Annu Rev Microbiol. 2009;63:61–78. doi: 10.1146/annurev.micro.091208.073210. [DOI] [PubMed] [Google Scholar]
- 8.Lee JW, Beebe K, Nangle LA, Jang J, Longo-Guess CM, Cook SA, Davisson MT, Sundberg JP, Schimmel P, Ackerman SL. Nature. 2006;443:50–55. doi: 10.1038/nature05096. [DOI] [PubMed] [Google Scholar]
- 9.Nangle LA, Motta CM, Schimmel P. Chem Biol. 2006;13:1091–1100. doi: 10.1016/j.chembiol.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 10.Jakubowski H. Acta Biochim Pol. 2011;58:149–163. [PubMed] [Google Scholar]
- 11.Martinis SA, Boniecki MT. FEBS Lett. 2010;584:455–459. doi: 10.1016/j.febslet.2009.11.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beuning PJ, Musier-Forsyth K. Proc Natl Acad Sci USA. 2000;97:8916–8920. doi: 10.1073/pnas.97.16.8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong FC, Beuning PJ, Nagan M, Shiba K, Musier-Forsyth K. Biochemistry. 2002;41:7108–7115. doi: 10.1021/bi012178j. [DOI] [PubMed] [Google Scholar]
- 14.An S, Musier-Forsyth K. J Biol Chem. 2005;280:34465–34472. doi: 10.1074/jbc.M507550200. [DOI] [PubMed] [Google Scholar]
- 15.Ruan B, Söll D. J Biol Chem. 2005;280:25887–25891. doi: 10.1074/jbc.M502174200. [DOI] [PubMed] [Google Scholar]
- 16.So BR, An S, Kumar S, Das M, Turner DA, Hadad CM, Musier-Forsyth K. J Biol Chem. 2011;286:31810–31820. doi: 10.1074/jbc.M111.232611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hussain T, Kruparani SP, Pal B, Dock-Bregeon AC, Dwivedi S, Shekar MR, Sureshbabu K, Sankaranarayanan R. EMBO J. 2006;25:4152–4162. doi: 10.1038/sj.emboj.7601278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nordin BE, Schimmel P. J Biol Chem. 2002;277:20510–20517. doi: 10.1074/jbc.M202023200. [DOI] [PubMed] [Google Scholar]
- 19.Ling J, Roy H, Ibba M. Proc Natl Acad Sci USA. 2007;104:72–77. doi: 10.1073/pnas.0606272104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boero M. J Phys Chem B. 2011;115:12276–12286. doi: 10.1021/jp2070024. [DOI] [PubMed] [Google Scholar]
- 21.Hagiwara Y, Field MJ, Nureki O, Tateno M. J Am Chem Soc. 2010;132:2751–2758. doi: 10.1021/ja9095208. [DOI] [PubMed] [Google Scholar]
- 22.Kumar S, Das M, Hadad CM, Musier-Forsyth K. J Biol Chem. 2012;287:3175–3184. doi: 10.1074/jbc.M111.313619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 24.Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, et al. J Comput Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 25.Case DA, Darden TA, Cheatham I, TE, Simmerling CL, Wang J, Duke RE, Luo R, Crowley M, Walker RC, Zhang W, et al. AMBER. Vol. 10. University of California; 2008. [Google Scholar]
- 26.Chemshell. A Computational Chemistry Shell. see www.chemshell.org.
- 27.Sherwood P, de Vries AH, Guest MF, Schreckenbach G, Catlow CRA, French SA, Sokol AA, Bromley ST, Thiel W, Turner AJ, et al. J Mol Struct. 2003;632:1–28. [Google Scholar]
- 28.TURBOMOLE V5-10 2008, a development of University of Karlsruhe and Forschungszentrum Karlsruhe GmBH. TURBOMOLE GmBH. 1989–2007 since 2007; available from http://www.turbomole.com.
- 29.Smith W, Forester TR. J Mol Graphics. 1996;14:136–141. doi: 10.1016/s0263-7855(96)00043-4. [DOI] [PubMed] [Google Scholar]
- 30.MacKerell AD, Bashford D, Bellott Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, et al. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 31.Becke AD. Phys Rev A. 1988;38:3098–3100. doi: 10.1103/physreva.38.3098. [DOI] [PubMed] [Google Scholar]
- 32.Perdew JP. Phys Rev B: Condens Matter. 1986;33:8822–8824. doi: 10.1103/physrevb.33.8822. [DOI] [PubMed] [Google Scholar]
- 33.Schäfer A, Horn H, Ahlrichs R. J Chem Phys. 1992;97:2571–2577. [Google Scholar]
- 34.Weigend F, Häser M, Patzelt H, Ahlrichs R. Chem Phys Lett. 1998;294:143–152. [Google Scholar]
- 35.Beebe K, de Pouplana LR, Schimmel P. EMBO J. 2003;22:668–675. doi: 10.1093/emboj/cdg065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bradford MM. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 37.Fersht AR, Ashford JS, Bruton CJ, Jakes R, Koch GLE, Hartley BS. Biochemistry. 1975;14:1–4. doi: 10.1021/bi00672a001. [DOI] [PubMed] [Google Scholar]
- 38.Beckman J, Kincaid K, Hocek M, Spratt T, Engels J, Cosstick R, Kuchta RD. Biochemistry. 2006;46:448–460. doi: 10.1021/bi061243s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu H, Kessler J, Peterson R, Musier-Forsyth K. Biochemistry. 1995;34:9795–9800. doi: 10.1021/bi00030a017. [DOI] [PubMed] [Google Scholar]
- 40.Minajigi A, Francklyn CS. Proc Natl Acad Sci USA. 2008;105:17748–17753. doi: 10.1073/pnas.0804247105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boyarshin KS, Kriklivyi IA, Rayevsky AV, Himin AA, Yaremchuk AD, Tukalo MA. Biopolymers & Cell. 2009;25:39–43. [Google Scholar]
- 42.Crepin T, Yaremchuk A, Tukalo M, Cusack S. Structure. 2006;14:1511–1525. doi: 10.1016/j.str.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 43.Zhang H, Huang K, Li Z, Banerjei L, Fisher KE, Grishin NV, Eisenstein E, Herzberg O. Proteins: Struct Funct Bioinform. 2000;40:86–97. doi: 10.1002/(sici)1097-0134(20000701)40:1<86::aid-prot100>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 44.Lim K, Tempczyk A, Bonander N, Toedt J, Howard A, Eisenstein E, Herzberg O. J Biol Chem. 2003;278:13496–13502. doi: 10.1074/jbc.M213150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fedor MJ. Annu Rev Biophys. 2009;38:271–299. doi: 10.1146/annurev.biophys.050708.133710. [DOI] [PubMed] [Google Scholar]
- 46.Wilcox JL, Ahluwalia AK, Bevilacqua PC. Acc Chem Res. 2011;44:1270–1279. doi: 10.1021/ar2000452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilson TJ, Li NS, Lu J, Frederiksen JK, Piccirilli JA, Lilley DMJ. Proc Natl Acad Sci USA. 2010;107:11751–11756. doi: 10.1073/pnas.1004255107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Grotthuss CJT. Anal Chim. 1806;58:54–73. [Google Scholar]
- 49.Cisneros GA, Wang M, Silinski P, Fitzgerald MC, Yang W. Biochemistry. 2004;43:6885–6892. doi: 10.1021/bi049943p. [DOI] [PubMed] [Google Scholar]
- 50.Dobson RCJ, Griffin MDW, Devenish SRA, Pearce FG, Hutton CA, Gerrard JA, Jameson GB, Perugini MA. Protein Sci. 2008;17:2080–2090. doi: 10.1110/ps.037440.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wymore T, Deerfield DW, Hempel J. Biochemistry. 2007;46:9495–9506. doi: 10.1021/bi700555g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cleland WW, Kreevoy MM. Science. 1994;264:1887–1890. doi: 10.1126/science.8009219. [DOI] [PubMed] [Google Scholar]
- 53.Tuckerman ME, Marx D, Klein ML, Parrinello M. Science. 1997;275:817–820. doi: 10.1126/science.275.5301.817. [DOI] [PubMed] [Google Scholar]
- 54.Zundel G, Metzger H. Z Phys Chem. 1968;58:225–245. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.