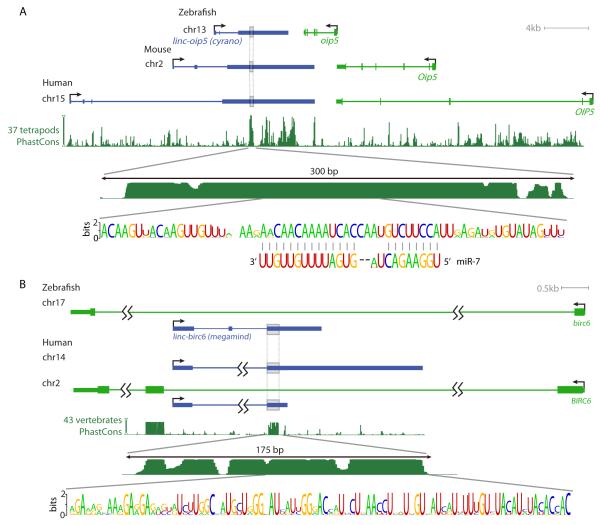
Figure 4. lincRNAs with Short Conserved Segments.
(A) Genomic context and sequence conservation of the linc-oip5 (cyrano) lincRNA gene. Gray boxes include the deeply conserved region. The conservation plot is relative to the human locus, and is based on aligned regions of 37 genomes, which do not include any fish genomes, as those do not contain any regions that are aligned with this human locus in the whole-genome alignments. The top consensus logo highlights the RNA sequence of the most conserved segment, which we identified in 45 vertebrate genomes, including fish genomes. Shown are the 67 aligned positions present in zebrafish, with a score of 2 bits indicating residues perfectly conserved in all 45 genomes. The bottom consensus logo shows conservation of vertebrate miR-7 sequences annotated in miRBase 18, with vertical lines indicating Watson-Crick base pairs.
(B) Genomic context and sequence conservation of the linc-birc6 (megamind) lincRNA gene. As in (A), except the region is aligned to fish genomes in the whole-genome alignments, and the consensus logo is for the RNA sequence inferred from 75 sequences from 47 vertebrate genomes. An alternative isoform of the zebrafish RNA retains the first intron (Figure S3E).