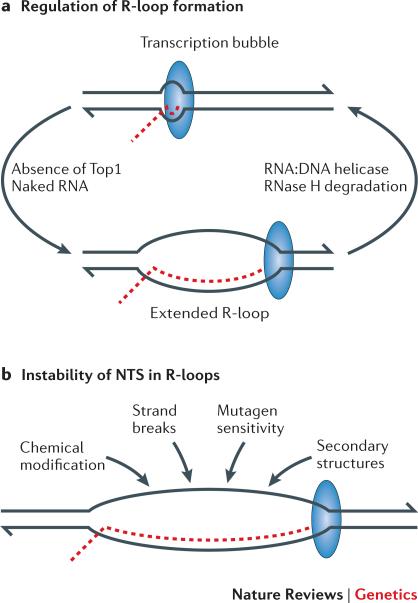
Figure 2. Factors that promote and remove R-loops during transcription.
a | R-loop formation is favored by the negative supercoils that accumulate in the absence of Top1 and by naked RNA that fails to be engaged immediately after transcription. In bacteria, the coupling between transcription and translation prevents the accumulation of naked RNA. In eukaryotes, RNA is co-transcriptionally assembled into ribonucleoprotein particles for splicing and/or nuclear transport. R-loops can be actively unwound by an RNA:DNA helicase or the RNA component degraded by RNase H. b | Factors expected to affect the exposed nontranscribed strand within R-loops. DNA strands are in black, with 3′ ends indicated by the half-arrowheads; the RNA transcript is in red; and the large blue oval corresponds to RNA polymerase.