Figure 3.
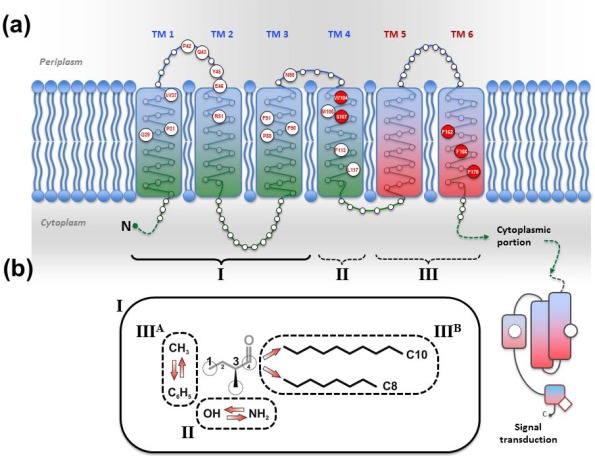
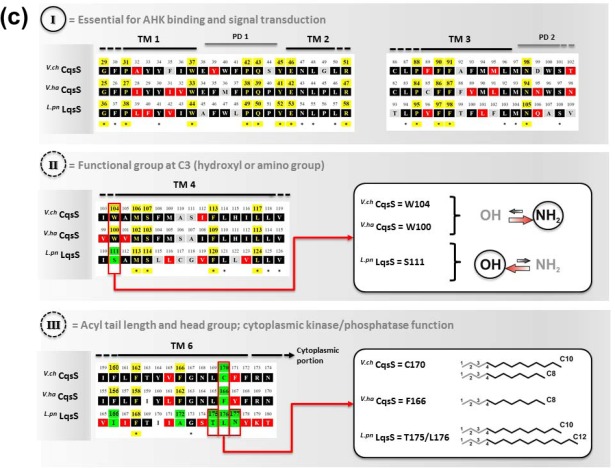
Signal recognition by the V. cholerae CqsS and L. pneumophila LqsS sensor kinases. (a) Topology model of the V. cholerae CqsS trans-membrane sensor domain based on the secondary structure prediction algorithms TMPRED and TMHMM. The N-terminal sensor domain encompasses six trans-membrane α-helices and defines the specificity and sensitivity for distinct AHK molecules (enlarged circles, amino acid position and one-letter code). (b) Sensor domain regions (I, II, III) define the interactions with functional groups of AHK ligands. (I) TM 1–3 and periplasmic domain PD 1 contain conserved amino acid clusters essential for ligand binding and signal transduction (cytoplasmic domain CD 1). (II) TM 4 (W104, S107) discriminates between hydroxyl and amino groups at C3 of AHK ligands. (III) TM 6 (F162, C170) determines binding to the head group (methyl vs. phenyl) and the acyl tail of AHK ligands. (c) Amino acid sequence alignment (Clustal Omega algortithm) of the sensor domains of V. cholerae (V.ch) CqsS, V. harveyi (V.ha) CqsS and L. pneumophila (L.pn) LqsS. The positions of species-specific amino acids in the sensor domain regions (I, II, III) are indicated, and stars denote conserved amino acids. Color scheme: black (2/3 conserved); red (similar amino acids); grey (different amino acids); yellow (amino acids defining ligand sensitivity, specificity and signal transduction motifs); green (amino acid exchange results in altered ligand specificity). Boxes illustrate altered ligand preferences due to site-specific amino acid polymorphisms in sensor domain region II and III.