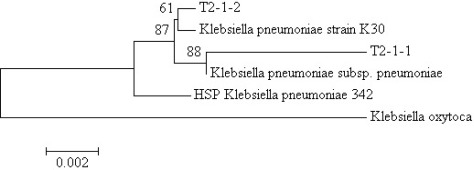
Figure 2.
16S rDNA-based phylogenetic tree showing the phylogenetic positions of oral bacterial isolates T2-1-1 and T2-1-2. A total of 912 unambiguously aligned nucleotides were analysed using MEGA 4. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The bar at the bottom represents evolutionary distance as 0.002 change per nucleotide position. GenBank accession numbers are given in parentheses: K. oxytoca (AB476819), K. pneumoniae subsp. pneumoniae (CP000647), K. pneumoniae strain K30 (EU661377), and K. pneumoniae 342 (CP000964).