Figure 5.
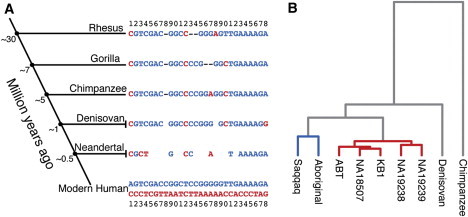
Evolution of FADS Haplotypes
(A) The 28 SNPs distinguishing the two main haplotypes in modern humans are shown at the bottom (haplotype A in red letters, D in blue), and the corresponding nucleotides in primates and archaic hominins are aligned above. The nucleotides for rhesus macaques, gorillas, and chimpanzees are taken from their respective reference genomes (rheMac2, gorGor3, and panTro2). Positions marked by hyphens are missing from the reference assemblies and probably represent deletions. For the archaic hominins, all nucleotides identified by at least ten reads (Denisovan) and two reads (Neanderthal) by Illumina sequencing are shown (see Table S2 for detailed data). Empty cells indicate positions with no sequence-read information as a result of either insufficient coverage or a deletion.
(B) Dendrogram—resulting from a hierarchical clustering via the UPGMA method—of pair-wise nucleotide differences in the FADS region between five DD individuals, two AA individuals, the Denovisan, and one chimpanzee. AA genotypes are depicted with blue branches, and DD genotypes are depicted with red branches.
